ADF Diagnostics In Jupyter#
This notebook will run the Atmospheric Diagnostic Framework using the settings in a config.yaml file in your ADF directory.
Note that it was developed to run on Cheyenne/Caspar with the NPL (conda) kernel
Setup#
Required packages#
import os.path
from pathlib import Path
import sys
Paths#
### default parameters
# adf_path = "../../externals/ADF"
# config_path = "."
# config_fil_str = "config_f.cam6_3_119.FLTHIST_ne30.r328_gamma0.33_soae_numcin3.001_vs_f.cam6_3_119.FLTHIST_ne30.r328_gamma0.33_soae.001.yaml"
# cell to insert parameters
# Parameters
test_global_param = "hello"
sname = "adf-quick-run"
adf_path = "../../externals/ADF"
config_path = "."
config_fil_str = "config_f.cam6_3_119.FLTHIST_ne30.r328_gamma0.33_soae.001.yaml"
subset_kwargs = {}
product = "/glade/u/home/hannay/CUPiD/examples/adf-mom6/computed_notebooks/adf-quick-run/adf_quick_run.ipynb"
All path processing:#
# Determine ADF directory path
# If it is in your cwd, set adf_path = local_path,
# otherwise set adf_path appropriately
local_path = os.path.abspath('')
# Set up the ADF for your location/user name
#user = "richling"
#adf_path = f"/glade/work/{user}/ADF/"
print(f"current working directory = {local_path}")
print(f"ADF path = {adf_path}")
current working directory = /glade/u/home/hannay/CUPiD/examples/nblibrary
ADF path = ../../externals/ADF
#set path to ADF lib
lib_path = os.path.join(adf_path,"lib")
print(f"The lib scripts live here, right? {lib_path}")
#Add paths to python path:
sys.path.append(lib_path)
The lib scripts live here, right? ../../externals/ADF/lib
#set path to ADF plotting scripts directory
plotting_scripts_path = os.path.join(adf_path,"scripts","plotting")
print(f"The plotting scripts live here, right? {plotting_scripts_path}")
#Add paths to python path:
sys.path.append(plotting_scripts_path)
The plotting scripts live here, right? ../../externals/ADF/scripts/plotting
Import config file into ADF object#
If there are errors, here, it is likely due to path errors above
config_file=os.path.join(config_path,config_fil_str)
#import ADF diagnostics object
from adf_diag import AdfDiag
# If this fails, check your paths output in the cells above,
# and that you are running the NPL (conda) Kernel
# You can see all the paths being examined by un-commenting the following:
#sys.path
#Initialize ADF object
adf = AdfDiag(config_file)
adf
<adf_diag.AdfDiag at 0x7f0d2469ebd0>
ADF Standard Work Flow#
Calculate the Time Series files#
NOTE: If not comparing against observations, you must run create_time_series() again with baseline flag
#Create model time series.
adf.create_time_series()
#Create model baseline time series (if needed):
if not adf.compare_obs:
adf.create_time_series(baseline=True)
Generating CAM time series files...
Processing time series for case 'b.e23_alpha16b.BLT1850.ne30_t232.054' :
WARNING: AODDUSTdn is not in the file /glade/campaign/cesm/development/cross-wg/diagnostic_framework/CESM_output_for_testing/b.e23_alpha16b.BLT1850.ne30_t232.054/atm/hist/b.e23_alpha16b.BLT1850.ne30_t232.054.cam.h0.0091-01.nc. No time series will be generated.
WARNING: TROP_T is not in the file /glade/campaign/cesm/development/cross-wg/diagnostic_framework/CESM_output_for_testing/b.e23_alpha16b.BLT1850.ne30_t232.054/atm/hist/b.e23_alpha16b.BLT1850.ne30_t232.054.cam.h0.0091-01.nc. No time series will be generated.
...CAM time series file generation has finished successfully.
Generating CAM time series files...
Processing time series for case 'f.cam6_3_119.FLTHIST_ne30.r328_gamma0.33_soae.001' :
WARNING: AODDUSTdn is not in the file /glade/campaign/cesm/development/cross-wg/diagnostic_framework/CESM_output_for_testing/f.cam6_3_119.FLTHIST_ne30.r328_gamma0.33_soae.001/atm/hist/f.cam6_3_119.FLTHIST_ne30.r328_gamma0.33_soae.001.cam.h0.1995-01.nc. No time series will be generated.
WARNING: TROP_T is not in the file /glade/campaign/cesm/development/cross-wg/diagnostic_framework/CESM_output_for_testing/f.cam6_3_119.FLTHIST_ne30.r328_gamma0.33_soae.001/atm/hist/f.cam6_3_119.FLTHIST_ne30.r328_gamma0.33_soae.001.cam.h0.1995-01.nc. No time series will be generated.
...CAM time series file generation has finished successfully.
Calculate the Climo files#
NOTE: Do not need to specify or repeat for baseline case unlike time series generation
#Create model climatology (climo) files.
adf.create_climo()
Calculating CAM climatologies...
Calculating climatologies for case 'b.e23_alpha16b.BLT1850.ne30_t232.054' :
The input location searched was: /glade/campaign/cesm/development/cross-wg/diagnostic_framework/CESM_output_for_testing/b.e23_alpha16b.BLT1850.ne30_t232.054/91-101. The glob pattern was b.e23_alpha16b.BLT1850.ne30_t232.054*.AODDUSTdn.*nc.
INFO: Found climo file and clobber is False, so skipping AODDUST and moving to next variable.
INFO: Found climo file and clobber is False, so skipping AODVIS and moving to next variable.
INFO: Found climo file and clobber is False, so skipping CLDHGH and moving to next variable.
INFO: Found climo file and clobber is False, so skipping CLDICE and moving to next variable.
INFO: Found climo file and clobber is False, so skipping CLDLIQ and moving to next variable.
INFO: Found climo file and clobber is False, so skipping CLDLOW and moving to next variable.
INFO: Found climo file and clobber is False, so skipping CLDMED and moving to next variable.
INFO: Found climo file and clobber is False, so skipping CLDTOT and moving to next variable.
INFO: Found climo file and clobber is False, so skipping CLOUD and moving to next variable.
INFO: Found climo file and clobber is False, so skipping FLNS and moving to next variable.
INFO: Found climo file and clobber is False, so skipping FLNT and moving to next variable.
INFO: Found climo file and clobber is False, so skipping FLNTC and moving to next variable.
INFO: Found climo file and clobber is False, so skipping FSNS and moving to next variable.
INFO: Found climo file and clobber is False, so skipping FSNT and moving to next variable.
INFO: Found climo file and clobber is False, so skipping FSNTC and moving to next variable.
INFO: Found climo file and clobber is False, so skipping LHFLX and moving to next variable.
INFO: Found climo file and clobber is False, so skipping LWCF and moving to next variable.
INFO: Found climo file and clobber is False, so skipping OMEGA500 and moving to next variable.
INFO: Found climo file and clobber is False, so skipping PBLH and moving to next variable.
INFO: Found climo file and clobber is False, so skipping PRECL and moving to next variable.
INFO: Found climo file and clobber is False, so skipping PRECT and moving to next variable.
INFO: Found climo file and clobber is False, so skipping PRECSL and moving to next variable.
INFO: Found climo file and clobber is False, so skipping PRECSC and moving to next variable.
INFO: Found climo file and clobber is False, so skipping PRECSC and moving to next variable.
INFO: Found climo file and clobber is False, so skipping PRECC and moving to next variable.
INFO: Found climo file and clobber is False, so skipping PS and moving to next variable.
INFO: Found climo file and clobber is False, so skipping PSL and moving to next variable.
INFO: Found climo file and clobber is False, so skipping QFLX and moving to next variable.
INFO: Found climo file and clobber is False, so skipping Q and moving to next variable.
INFO: Found climo file and clobber is False, so skipping RELHUM and moving to next variable.
INFO: Found climo file and clobber is False, so skipping SHFLX and moving to next variable.
INFO: Found climo file and clobber is False, so skipping SST and moving to next variable.
INFO: Found climo file and clobber is False, so skipping SWCF and moving to next variable.
INFO: Found climo file and clobber is False, so skipping T and moving to next variable.
INFO: Found climo file and clobber is False, so skipping TAUX and moving to next variable.
INFO: Found climo file and clobber is False, so skipping TAUY and moving to next variable.
INFO: Found climo file and clobber is False, so skipping TGCLDIWP and moving to next variable.
INFO: Found climo file and clobber is False, so skipping TGCLDLWP and moving to next variable.
INFO: Found climo file and clobber is False, so skipping TMQ and moving to next variable.
INFO: Found climo file and clobber is False, so skipping TREFHT and moving to next variable.
The input location searched was: /glade/campaign/cesm/development/cross-wg/diagnostic_framework/CESM_output_for_testing/b.e23_alpha16b.BLT1850.ne30_t232.054/91-101. The glob pattern was b.e23_alpha16b.BLT1850.ne30_t232.054*.TROP_T.*nc.
INFO: Found climo file and clobber is False, so skipping TS and moving to next variable.
INFO: Found climo file and clobber is False, so skipping U and moving to next variable.
INFO: Found climo file and clobber is False, so skipping U10 and moving to next variable.
INFO: Found climo file and clobber is False, so skipping ICEFRAC and moving to next variable.
INFO: Found climo file and clobber is False, so skipping OCNFRAC and moving to next variable.
INFO: Found climo file and clobber is False, so skipping LANDFRAC and moving to next variable.
Time series files for variable 'AODDUSTdn' not found. Script will continue to next variable.
Time series files for variable 'TROP_T' not found. Script will continue to next variable.
Calculating climatologies for case 'f.cam6_3_119.FLTHIST_ne30.r328_gamma0.33_soae.001' :
The input location searched was: /glade/campaign/cesm/development/cross-wg/diagnostic_framework/ADF-data/timeseries/f.cam6_3_119.FLTHIST_ne30.r328_gamma0.33_soae.001/1995-2005. The glob pattern was f.cam6_3_119.FLTHIST_ne30.r328_gamma0.33_soae.001*.AODDUSTdn.*nc.
INFO: Found climo file and clobber is False, so skipping AODDUST and moving to next variable.
INFO: Found climo file and clobber is False, so skipping AODVIS and moving to next variable.
INFO: Found climo file and clobber is False, so skipping CLDHGH and moving to next variable.
INFO: Found climo file and clobber is False, so skipping CLDICE and moving to next variable.
INFO: Found climo file and clobber is False, so skipping CLDLIQ and moving to next variable.
INFO: Found climo file and clobber is False, so skipping CLDLOW and moving to next variable.
INFO: Found climo file and clobber is False, so skipping CLDMED and moving to next variable.
INFO: Found climo file and clobber is False, so skipping CLDTOT and moving to next variable.
INFO: Found climo file and clobber is False, so skipping CLOUD and moving to next variable.
INFO: Found climo file and clobber is False, so skipping FLNS and moving to next variable.
INFO: Found climo file and clobber is False, so skipping FLNT and moving to next variable.
INFO: Found climo file and clobber is False, so skipping FLNTC and moving to next variable.
INFO: Found climo file and clobber is False, so skipping FSNS and moving to next variable.
INFO: Found climo file and clobber is False, so skipping FSNT and moving to next variable.
INFO: Found climo file and clobber is False, so skipping FSNTC and moving to next variable.
INFO: Found climo file and clobber is False, so skipping LHFLX and moving to next variable.
INFO: Found climo file and clobber is False, so skipping LWCF and moving to next variable.
INFO: Found climo file and clobber is False, so skipping OMEGA500 and moving to next variable.
INFO: Found climo file and clobber is False, so skipping PBLH and moving to next variable.
INFO: Found climo file and clobber is False, so skipping PRECL and moving to next variable.
INFO: Found climo file and clobber is False, so skipping PRECT and moving to next variable.
INFO: Found climo file and clobber is False, so skipping PRECSL and moving to next variable.
INFO: Found climo file and clobber is False, so skipping PRECSC and moving to next variable.
INFO: Found climo file and clobber is False, so skipping PRECSC and moving to next variable.
INFO: Found climo file and clobber is False, so skipping PRECC and moving to next variable.
INFO: Found climo file and clobber is False, so skipping PS and moving to next variable.
INFO: Found climo file and clobber is False, so skipping PSL and moving to next variable.
INFO: Found climo file and clobber is False, so skipping QFLX and moving to next variable.
INFO: Found climo file and clobber is False, so skipping Q and moving to next variable.
INFO: Found climo file and clobber is False, so skipping RELHUM and moving to next variable.
INFO: Found climo file and clobber is False, so skipping SHFLX and moving to next variable.
INFO: Found climo file and clobber is False, so skipping SST and moving to next variable.
INFO: Found climo file and clobber is False, so skipping SWCF and moving to next variable.
INFO: Found climo file and clobber is False, so skipping T and moving to next variable.
INFO: Found climo file and clobber is False, so skipping TAUX and moving to next variable.
INFO: Found climo file and clobber is False, so skipping TAUY and moving to next variable.
INFO: Found climo file and clobber is False, so skipping TGCLDIWP and moving to next variable.
INFO: Found climo file and clobber is False, so skipping TGCLDLWP and moving to next variable.
INFO: Found climo file and clobber is False, so skipping TMQ and moving to next variable.
INFO: Found climo file and clobber is False, so skipping TREFHT and moving to next variable.
The input location searched was: /glade/campaign/cesm/development/cross-wg/diagnostic_framework/ADF-data/timeseries/f.cam6_3_119.FLTHIST_ne30.r328_gamma0.33_soae.001/1995-2005. The glob pattern was f.cam6_3_119.FLTHIST_ne30.r328_gamma0.33_soae.001*.TROP_T.*nc.
INFO: Found climo file and clobber is False, so skipping TS and moving to next variable.
INFO: Found climo file and clobber is False, so skipping U and moving to next variable.
INFO: Found climo file and clobber is False, so skipping U10 and moving to next variable.
INFO: Found climo file and clobber is False, so skipping ICEFRAC and moving to next variable.
INFO: Found climo file and clobber is False, so skipping OCNFRAC and moving to next variable.
INFO: Found climo file and clobber is False, so skipping LANDFRAC and moving to next variable.
...CAM climatologies have been calculated successfully.
Generating CAM TEM diagnostics files...
Processing TEM for case 'b.e23_alpha16b.BLT1850.ne30_t232.054' :
INFO: Found TEM file and clobber is False, so moving to next case.
Processing TEM for case 'f.cam6_3_119.FLTHIST_ne30.r328_gamma0.33_soae.001' :
INFO: Found TEM file and clobber is False, so moving to next case.
...TEM variables have been calculated successfully.
Regrid the Climo files#
#Regrid model climatology files to match either
#observations or CAM baseline climatologies.
#This call uses the "regridding_scripts" specified in the config file:
adf.regrid_climo()
Regridding CAM climatologies...
Regridding case 'b.e23_alpha16b.BLT1850.ne30_t232.054' :
- regridding PS (known targets: ['f.cam6_3_119.FLTHIST_ne30.r328_gamma0.33_soae.001'])
Regridded file already exists, so skipping...
- regridding LANDFRAC (known targets: ['f.cam6_3_119.FLTHIST_ne30.r328_gamma0.33_soae.001'])
Regridded file already exists, so skipping...
- regridding OCNFRAC (known targets: ['f.cam6_3_119.FLTHIST_ne30.r328_gamma0.33_soae.001'])
Regridded file already exists, so skipping...
- regridding AODDUSTdn (known targets: ['f.cam6_3_119.FLTHIST_ne30.r328_gamma0.33_soae.001'])
- regridding AODDUSTdn failed, no file. Continuing to next variable.
- regridding AODDUST (known targets: ['f.cam6_3_119.FLTHIST_ne30.r328_gamma0.33_soae.001'])
Regridded file already exists, so skipping...
- regridding AODVIS (known targets: ['f.cam6_3_119.FLTHIST_ne30.r328_gamma0.33_soae.001'])
Regridded file already exists, so skipping...
- regridding CLDHGH (known targets: ['f.cam6_3_119.FLTHIST_ne30.r328_gamma0.33_soae.001'])
Regridded file already exists, so skipping...
- regridding CLDICE (known targets: ['f.cam6_3_119.FLTHIST_ne30.r328_gamma0.33_soae.001'])
Regridded file already exists, so skipping...
- regridding CLDLIQ (known targets: ['f.cam6_3_119.FLTHIST_ne30.r328_gamma0.33_soae.001'])
Regridded file already exists, so skipping...
- regridding CLDLOW (known targets: ['f.cam6_3_119.FLTHIST_ne30.r328_gamma0.33_soae.001'])
Regridded file already exists, so skipping...
- regridding CLDMED (known targets: ['f.cam6_3_119.FLTHIST_ne30.r328_gamma0.33_soae.001'])
Regridded file already exists, so skipping...
- regridding CLDTOT (known targets: ['f.cam6_3_119.FLTHIST_ne30.r328_gamma0.33_soae.001'])
Regridded file already exists, so skipping...
- regridding CLOUD (known targets: ['f.cam6_3_119.FLTHIST_ne30.r328_gamma0.33_soae.001'])
Regridded file already exists, so skipping...
- regridding FLNS (known targets: ['f.cam6_3_119.FLTHIST_ne30.r328_gamma0.33_soae.001'])
Regridded file already exists, so skipping...
- regridding FLNT (known targets: ['f.cam6_3_119.FLTHIST_ne30.r328_gamma0.33_soae.001'])
Regridded file already exists, so skipping...
- regridding FLNTC (known targets: ['f.cam6_3_119.FLTHIST_ne30.r328_gamma0.33_soae.001'])
Regridded file already exists, so skipping...
- regridding FSNS (known targets: ['f.cam6_3_119.FLTHIST_ne30.r328_gamma0.33_soae.001'])
Regridded file already exists, so skipping...
- regridding FSNT (known targets: ['f.cam6_3_119.FLTHIST_ne30.r328_gamma0.33_soae.001'])
Regridded file already exists, so skipping...
- regridding FSNTC (known targets: ['f.cam6_3_119.FLTHIST_ne30.r328_gamma0.33_soae.001'])
Regridded file already exists, so skipping...
- regridding LHFLX (known targets: ['f.cam6_3_119.FLTHIST_ne30.r328_gamma0.33_soae.001'])
Regridded file already exists, so skipping...
- regridding LWCF (known targets: ['f.cam6_3_119.FLTHIST_ne30.r328_gamma0.33_soae.001'])
Regridded file already exists, so skipping...
- regridding OMEGA500 (known targets: ['f.cam6_3_119.FLTHIST_ne30.r328_gamma0.33_soae.001'])
Regridded file already exists, so skipping...
- regridding PBLH (known targets: ['f.cam6_3_119.FLTHIST_ne30.r328_gamma0.33_soae.001'])
Regridded file already exists, so skipping...
- regridding PRECL (known targets: ['f.cam6_3_119.FLTHIST_ne30.r328_gamma0.33_soae.001'])
Regridded file already exists, so skipping...
- regridding PRECT (known targets: ['f.cam6_3_119.FLTHIST_ne30.r328_gamma0.33_soae.001'])
Regridded file already exists, so skipping...
- regridding PRECSL (known targets: ['f.cam6_3_119.FLTHIST_ne30.r328_gamma0.33_soae.001'])
Regridded file already exists, so skipping...
- regridding PRECSC (known targets: ['f.cam6_3_119.FLTHIST_ne30.r328_gamma0.33_soae.001'])
Regridded file already exists, so skipping...
- regridding PRECSC (known targets: ['f.cam6_3_119.FLTHIST_ne30.r328_gamma0.33_soae.001'])
Regridded file already exists, so skipping...
- regridding PRECC (known targets: ['f.cam6_3_119.FLTHIST_ne30.r328_gamma0.33_soae.001'])
Regridded file already exists, so skipping...
- regridding PSL (known targets: ['f.cam6_3_119.FLTHIST_ne30.r328_gamma0.33_soae.001'])
Regridded file already exists, so skipping...
- regridding QFLX (known targets: ['f.cam6_3_119.FLTHIST_ne30.r328_gamma0.33_soae.001'])
Regridded file already exists, so skipping...
- regridding Q (known targets: ['f.cam6_3_119.FLTHIST_ne30.r328_gamma0.33_soae.001'])
Regridded file already exists, so skipping...
- regridding RELHUM (known targets: ['f.cam6_3_119.FLTHIST_ne30.r328_gamma0.33_soae.001'])
Regridded file already exists, so skipping...
- regridding SHFLX (known targets: ['f.cam6_3_119.FLTHIST_ne30.r328_gamma0.33_soae.001'])
Regridded file already exists, so skipping...
- regridding SST (known targets: ['f.cam6_3_119.FLTHIST_ne30.r328_gamma0.33_soae.001'])
Regridded file already exists, so skipping...
- regridding SWCF (known targets: ['f.cam6_3_119.FLTHIST_ne30.r328_gamma0.33_soae.001'])
Regridded file already exists, so skipping...
- regridding T (known targets: ['f.cam6_3_119.FLTHIST_ne30.r328_gamma0.33_soae.001'])
Regridded file already exists, so skipping...
- regridding TAUX (known targets: ['f.cam6_3_119.FLTHIST_ne30.r328_gamma0.33_soae.001'])
Regridded file already exists, so skipping...
- regridding TAUY (known targets: ['f.cam6_3_119.FLTHIST_ne30.r328_gamma0.33_soae.001'])
Regridded file already exists, so skipping...
- regridding TGCLDIWP (known targets: ['f.cam6_3_119.FLTHIST_ne30.r328_gamma0.33_soae.001'])
Regridded file already exists, so skipping...
- regridding TGCLDLWP (known targets: ['f.cam6_3_119.FLTHIST_ne30.r328_gamma0.33_soae.001'])
Regridded file already exists, so skipping...
- regridding TMQ (known targets: ['f.cam6_3_119.FLTHIST_ne30.r328_gamma0.33_soae.001'])
Regridded file already exists, so skipping...
- regridding TREFHT (known targets: ['f.cam6_3_119.FLTHIST_ne30.r328_gamma0.33_soae.001'])
Regridded file already exists, so skipping...
- regridding TROP_T (known targets: ['f.cam6_3_119.FLTHIST_ne30.r328_gamma0.33_soae.001'])
- regridding TROP_T failed, no file. Continuing to next variable.
- regridding TS (known targets: ['f.cam6_3_119.FLTHIST_ne30.r328_gamma0.33_soae.001'])
Regridded file already exists, so skipping...
- regridding U (known targets: ['f.cam6_3_119.FLTHIST_ne30.r328_gamma0.33_soae.001'])
Regridded file already exists, so skipping...
- regridding U10 (known targets: ['f.cam6_3_119.FLTHIST_ne30.r328_gamma0.33_soae.001'])
Regridded file already exists, so skipping...
- regridding ICEFRAC (known targets: ['f.cam6_3_119.FLTHIST_ne30.r328_gamma0.33_soae.001'])
Regridded file already exists, so skipping...
...CAM climatologies have been regridded successfully.
Run statistics on Time Series files#
#Perform analyses on the simulation(s). #This call uses the “analysis_scripts” specified in the config file: adf.perform_analyses()
Create Plots#
#Create plots. #This call uses the “plotting_scripts” specified in the config file: adf.create_plots()
Generate HTML files#
This will create html files that you can view via webbrower either:
in Casper/Cheyenne
pushing it to CGD projects webpage through Tungsten
#Create website. if adf.create_html: adf.create_website()
ADF Helpful Methods and Structures#
Demonstration of a few methods to get information from the ADF object
Get information from the subsections of the config yaml file#
Remember the different sub-sections?
Basic Info Section#
basic_info_dict = adf.read_config_var("diag_basic_info")
for key,val in basic_info_dict.items():
print(f"{key}: {val}")
compare_obs: False
hist_str: cam.h0
create_html: True
obs_data_loc: /glade/work/nusbaume/SE_projects/model_diagnostics/ADF_obs
cam_regrid_loc: ${diag_loc}regrid/
cam_overwrite_regrid: False
cam_diag_plot_loc: ${diag_loc}diag-plot/
use_defaults: True
plot_press_levels: [200, 850]
weight_season: True
num_procs: 8
redo_plot: False
Test Case Info Section#
test_dict = adf.read_config_var("diag_cam_climo")
for key,val in test_dict.items():
print(f"{key}: {val}")
calc_cam_climo: True
cam_overwrite_climo: False
cam_case_name: b.e23_alpha16b.BLT1850.ne30_t232.054
cam_hist_loc: /glade/campaign/cesm/development/cross-wg/diagnostic_framework/CESM_output_for_testing/${diag_cam_climo.cam_case_name}/atm/hist/
yrs: ${diag_cam_climo.start_year}-${diag_cam_climo.end_year}
cam_climo_loc: ${climo_loc}${diag_cam_climo.cam_case_name}/atm/proc/${diag_cam_climo.yrs}/
start_year: 91
end_year: 101
cam_ts_done: False
cam_ts_save: True
cam_overwrite_ts: False
cam_ts_loc: ${ts_loc}${diag_cam_climo.cam_case_name}/${diag_cam_climo.yrs}/
case_nickname: ${diag_cam_climo.cam_case_name}
Baseline Case Info Section#
baseline_dict = adf.read_config_var("diag_cam_baseline_climo")
for key,val in baseline_dict.items():
print(f"{key}: {val}")
calc_cam_climo: True
cam_overwrite_climo: False
cam_case_name: f.cam6_3_119.FLTHIST_ne30.r328_gamma0.33_soae.001
cam_hist_loc: /glade/campaign/cesm/development/cross-wg/diagnostic_framework/CESM_output_for_testing/${diag_cam_baseline_climo.cam_case_name}/atm/hist/
yrs: ${diag_cam_baseline_climo.start_year}-${diag_cam_baseline_climo.end_year}
cam_climo_loc: /glade/campaign/cesm/development/cross-wg/diagnostic_framework/ADF-data/climos/${diag_cam_baseline_climo.cam_case_name}/${diag_cam_baseline_climo.yrs}/
start_year: 1995
end_year: 2005
cam_ts_done: False
cam_ts_save: True
cam_overwrite_ts: False
cam_ts_loc: /glade/campaign/cesm/development/cross-wg/diagnostic_framework/ADF-data/timeseries/${diag_cam_baseline_climo.cam_case_name}/${diag_cam_baseline_climo.yrs}/
case_nickname: ${diag_cam_baseline_climo.cam_case_name}
Get information not directly from the subsections of the config yaml file#
This just represents a different way to get some ADF info
Get Case/Baseline Names#
This is a different wat to get case names than from the adf.read_config_var() method that read in data from sub-sections above
#List of case names (list by default)
case_names = adf.get_cam_info("cam_case_name",required=True)
print(case_names)
base_name = adf.get_baseline_info("cam_case_name")
print(base_name)
['b.e23_alpha16b.BLT1850.ne30_t232.054']
f.cam6_3_119.FLTHIST_ne30.r328_gamma0.33_soae.001
Get Case/Baseline Climo file locations#
Here we are calling directly from the config file, no subsection
case_climo_loc = adf.get_cam_info('cam_climo_loc', required=True)
base_climo_loc = adf.get_baseline_info("cam_climo_loc")
case_climo_loc,base_climo_loc
(['/glade/campaign/cesm/development/cross-wg/diagnostic_framework/CESM_output_for_testing/b.e23_alpha16b.BLT1850.ne30_t232.054/atm/proc/91-101/'],
'/glade/campaign/cesm/development/cross-wg/diagnostic_framework/ADF-data/climos/f.cam6_3_119.FLTHIST_ne30.r328_gamma0.33_soae.001/1995-2005/')
Get Desired Variable Names#
Here we are calling directly from the config file, no subsection
var_list = adf.diag_var_list
print(var_list)
['AODDUSTdn', 'AODDUST', 'AODVIS', 'CLDHGH', 'CLDICE', 'CLDLIQ', 'CLDLOW', 'CLDMED', 'CLDTOT', 'CLOUD', 'FLNS', 'FLNT', 'FLNTC', 'FSNS', 'FSNT', 'FSNTC', 'LHFLX', 'LWCF', 'OMEGA500', 'PBLH', 'PRECL', 'PRECT', 'PRECSL', 'PRECSC', 'PRECSC', 'PRECC', 'PS', 'PSL', 'QFLX', 'Q', 'RELHUM', 'SHFLX', 'SST', 'SWCF', 'T', 'TAUX', 'TAUY', 'TGCLDIWP', 'TGCLDLWP', 'TMQ', 'TREFHT', 'TROP_T', 'TS', 'U', 'U10', 'ICEFRAC', 'OCNFRAC', 'LANDFRAC']
Get variable defaults from adf_variable_defaults.yaml#
Take a look at what defaults are for TS
adf.variable_defaults["TS"]
{'colormap': 'Blues',
'contour_levels_range': [220, 320, 5],
'diff_colormap': 'BrBG',
'diff_contour_range': [-10, 10, 1],
'scale_factor': 1,
'add_offset': 0,
'new_unit': 'K',
'mpl': {'colorbar': {'label': 'K'}},
'obs_file': 'ERAI_all_climo.nc',
'obs_name': 'ERAI',
'obs_var_name': 'TS',
'category': 'Surface variables'}
Exploration of the Output Data#
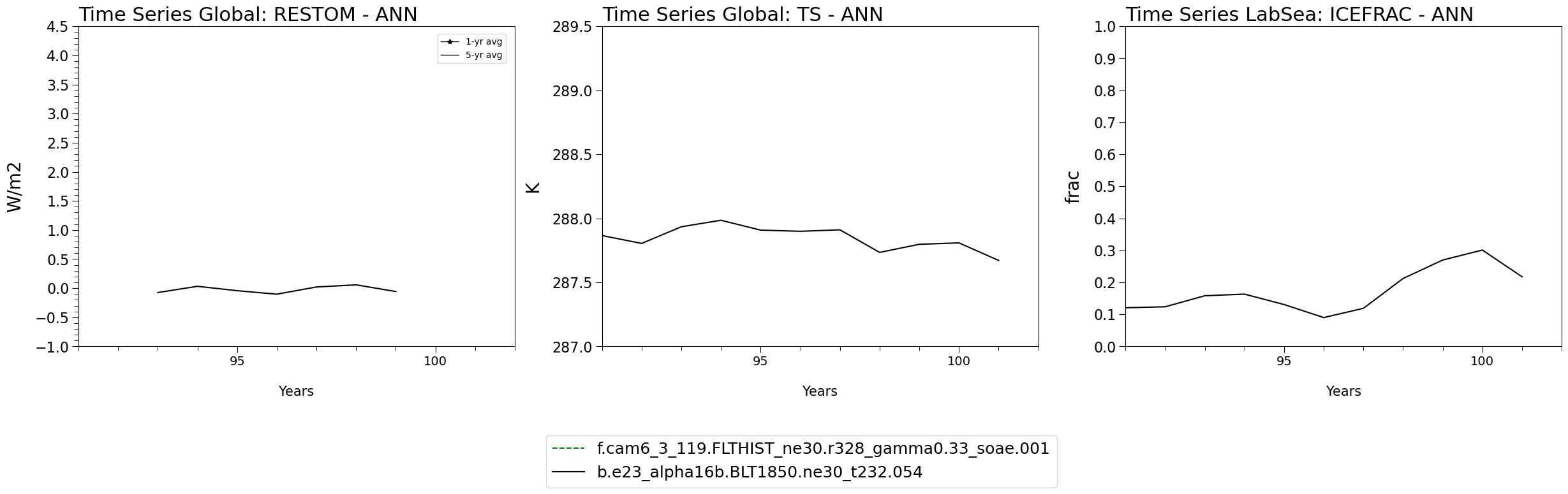
Now that the ADF has created all the necessary timeseries/climo/regridded data let’s run a quick set of functions to plot time series for RESTOM, TS, and ICEFRAC
Let’s grab the case names, time series locations, variable defaults dictionary and climo years#
case_names = adf.get_cam_info('cam_case_name', required=True)
case_names_len = len(case_names)
data_name = adf.get_baseline_info('cam_case_name', required=False)
case_ts_locs = adf.get_cam_info("cam_ts_loc", required=True)
data_ts_loc = adf.get_baseline_info("cam_ts_loc", required=False)
res = adf.variable_defaults # dict of variable-specific plot preferences
# or an empty dictionary if use_defaults was not specified in YAML.
start_year = adf.climo_yrs["syears"]
end_year = adf.climo_yrs["eyears"]
Time Series Plotting Functions#
def _load_dataset(fils):
if len(fils) == 0:
print("Input file list is empty.")
return None
elif len(fils) > 1:
return xr.open_mfdataset(fils, combine='by_coords')
else:
sfil = str(fils[0])
return xr.open_dataset(sfil)
#End if
#End def
def _data_calcs(ts_loc,var,subset=None):
"""
args
----
- ts_loc: Path
path to time series file
- var: str
name of variable
- subset (optional): dict
lat/lon extents (south, north, east, west)
"""
fils = sorted(list(Path(ts_loc).glob(f"*{var}*.nc")))
ts_ds = _load_dataset(fils)
time = ts_ds['time']
time = xr.DataArray(ts_ds['time_bnds'].load().mean(dim='nbnd').values, dims=time.dims, attrs=time.attrs)
ts_ds['time'] = time
ts_ds.assign_coords(time=time)
ts_ds = xr.decode_cf(ts_ds)
if subset != None:
ts_ds = ts_ds.sel(lat=slice(subset["s"],subset["n"]), lon=slice(subset["w"],subset["e"]))
data = ts_ds[var].squeeze()
unit = data.units
# global weighting
w = np.cos(np.radians(data.lat))
avg = data.weighted(w).mean(dim=("lat","lon"))
yrs = np.unique([str(val.item().timetuple().tm_year).zfill(4) for _,val in enumerate(ts_ds["time"])])
return avg,yrs,unit
def ts_plot(ax, name, vals, yrs, unit, color_dict,linewidth=None,zorder=1):
"""
args
----
- color_dict: dict
color and marker style for variable
"""
ax.plot(yrs, vals, color_dict["marker"], c=color_dict["color"],label=name,linewidth=linewidth,zorder=zorder)
ax.set_xlabel("Years",fontsize=15,labelpad=20)
ax.set_ylabel(unit,fontsize=15,labelpad=20)
# For the minor ticks, use no labels; default NullFormatter.
ax.tick_params(which='major', length=7)
ax.tick_params(which='minor', length=5)
return ax
def plot_var_details(ax, var, vals_cases, vals_base):
mins = []
maxs = []
for i,val in enumerate(vals_cases):
mins.append(np.nanmin(vals_cases[i]))
maxs.append(np.nanmax(vals_cases[i]))
mins.append(np.nanmin(vals_base))
maxs.append(np.nanmax(vals_base))
if var == "SST":
ax.set_ylabel("K",fontsize=20,labelpad=12)
tick_spacing = 0.5
ax.yaxis.set_major_locator(MultipleLocator(1))
ax.set_title(f"Time Series Global: {var} - ANN",loc="left",fontsize=22)
if var == "TS":
ax.set_ylabel("K",fontsize=20,labelpad=12)
tick_spacing = 0.5
ax.yaxis.set_minor_locator(MultipleLocator(0.5))
ax.set_title(f"Time Series Global: {var} - ANN",loc="left",fontsize=22)
if var == "ICEFRAC":
ax.set_ylabel("frac",fontsize=20,labelpad=12)
tick_spacing = 0.1
ax.set_ylim(np.floor(min(mins)),np.ceil(max(maxs)))
ax.set_title(f"Time Series LabSea: {var} - ANN",loc="left",fontsize=22)
if var == "RESTOM":
ax.set_ylabel("W/m2",fontsize=20,labelpad=12)
tick_spacing = 0.5
ax.yaxis.set_minor_locator(MultipleLocator(0.1))
ax.set_title(f"Time Series Global: {var} - ANN",loc="left",fontsize=22)
# Set label to show if RESTOM is 1 or 5-yr avg
line_1yr = Line2D([], [], label='1-yr avg', color='k', linewidth=1,marker='*',)
line_5yr = Line2D([], [], label='5-yr avg', color='k', linewidth=1,)
ax.legend(handles=[line_1yr,line_5yr], bbox_to_anchor=(0.99, 0.99))
# Add extra space on the y-axis, except for ICEFRAC
if var != "ICEFRAC":
ax.set_ylim(np.floor(min(mins)),np.ceil(max(maxs))+tick_spacing)
ax.yaxis.set_major_locator(MultipleLocator(tick_spacing))
ax.tick_params(axis='y', which='major', labelsize=16)
ax.tick_params(axis='y', which='minor', labelsize=16)
ax.tick_params(axis='x', which='major', labelsize=14)
ax.tick_params(axis='x', which='minor', labelsize=14)
return ax
Plot the time series!#
ts_var_list = ["RESTOM","TS","ICEFRAC"]
%matplotlib inline
import matplotlib.pyplot as plt
import xarray as xr
import numpy as np
import matplotlib.ticker as ticker
from matplotlib.ticker import MultipleLocator
from matplotlib.lines import Line2D
fig = plt.figure(figsize=(30,15))
# Change the layout/number of subplots based off number of variables desired
rows = 2
cols = 3
gs = fig.add_gridspec(rows, cols, hspace=.3, wspace=.2)
# Rough subset for Lab Sea
w = -63.5+360
e = -47.5+360
s = 53.5
n = 65.5
subset = {"s":s,"n":n,"e":e,"w":w}
# Add more colors as needed for number of test cases
# ** Baseline is already added as green dashed line in plotting function **
# matplotlib colors here: https://matplotlib.org/stable/gallery/color/named_colors.html
colors = ["k", "aqua", "orange", "b", "magenta", "goldenrod", "slategrey", "rosybrown"]
# Setup plotting
#---------------
# Loop over variables:
for i,var in enumerate(ts_var_list):
print("Plotting variable:",var)
if var == "RESTOM":
ax = plt.subplot(gs[0, 0])
if var == "TS":
ax = plt.subplot(gs[0, 1])
if var == "ICEFRAC":
ax = plt.subplot(gs[0, 2])
# Grab baseline case:
#--------------------
if var == "RESTOM":
avg_base_FSNT,yrs_base,unit = _data_calcs(data_ts_loc,'FSNT')
avg_base_FLNT,_,_ = _data_calcs(data_ts_loc,"FLNT")
if len(yrs_base) < 5:
print(f"Not a lot of climo years for {data_name}, only doing 1-yr avg for RESTOM...")
FSNT_base = avg_base_FSNT
FLNT_base = avg_base_FLNT
else:
FSNT_base = avg_base_FSNT.rolling(time=60,center=True).mean()
FLNT_base = avg_base_FLNT.rolling(time=60,center=True).mean()
avg_base = FSNT_base - FLNT_base
if (var == "TS" or var == "SST"):
avg_base,yrs_base,unit = _data_calcs(data_ts_loc,var)
if var == "ICEFRAC":
avg_base,yrs_base,unit = _data_calcs(data_ts_loc,var,subset)
# Get int of years for plotting on x-axis
yrs_base_int = yrs_base.astype(int)
# Create yearly averages
vals_base = [avg_base.sel(time=i).mean() for i in yrs_base]
# Plot baseline data
color_dict = {"color":"g","marker":"--"}
ax = ts_plot(ax, data_name, vals_base, yrs_base_int, unit, color_dict)
# Loop over test cases:
#----------------------
# Create lists to hold all sets of years (for each case) and
# sets of var data (for each case)
vals_cases = []
yrs_cases = []
for case_idx, case_name in enumerate(case_names):
if var == "RESTOM":
avg_case_FSNT,yrs_case,unit = _data_calcs(case_ts_locs[case_idx],'FSNT')
avg_case_FLNT,_,_ = _data_calcs(case_ts_locs[case_idx],"FLNT")
if len(yrs_case) < 5:
print(f"Not a lot of climo years for {case_name}, only doing 1-yr avg for RESTOM...")
FSNT_case = avg_case_FSNT
FLNT_case = avg_case_FLNT
color_dict = {"color":colors[case_idx],"marker":"-*"}
else:
FSNT_case = avg_case_FSNT.rolling(time=60,center=True).mean()
FLNT_case = avg_case_FLNT.rolling(time=60,center=True).mean()
color_dict = {"color":colors[case_idx],"marker":"-"}
avg_case = FSNT_case - FLNT_case
if var == "TS":
avg_case,yrs_case,unit = _data_calcs(case_ts_locs[case_idx],var)
color_dict = {"color":colors[case_idx],"marker":"-"}
if var == "ICEFRAC":
avg_case,yrs_case,unit = _data_calcs(case_ts_locs[case_idx],var,subset)
color_dict = {"color":colors[case_idx],"marker":"-"}
# Get yearly averages for all available years
vals_case = [avg_case.sel(time=i).mean() for i in yrs_case]
vals_cases.append(vals_case)
# Get int of years for plotting on x-axis
yrs_case_int = yrs_case.astype(int)
yrs_cases.append(yrs_case_int)
# Add case to plot (ax)
ax = ts_plot(ax, case_name, vals_case, yrs_case_int, unit, color_dict)
# End for (case names)
# Get variable details
ax = plot_var_details(ax, var, vals_cases, vals_base)
#Grab all unique years and find min/max years
uniq_yrs = sorted(x for v in yrs_cases for x in v)
max_year = int(max(uniq_yrs))
min_year = int(min(uniq_yrs))
last_year = max_year - max_year % 5
if (max_year > 5) and (last_year < max_year):
last_year += 5
first_year = min_year - min_year % 5
if min_year < 5:
first_year = 0
ax.set_xlim(first_year, last_year)
ax.set_xlabel("Years",fontsize=15,labelpad=20)
# Set the x-axis plot limits
# to guarantee data from all cases (including baseline) are on plot
ax.set_xlim(min_year, max_year+1)
# x-axis ticks and numbers
if max_year-min_year > 120:
ax.xaxis.set_major_locator(MultipleLocator(20))
ax.xaxis.set_minor_locator(MultipleLocator(10))
if 10 <= max_year-min_year <= 120:
ax.xaxis.set_major_locator(MultipleLocator(5))
ax.xaxis.set_minor_locator(MultipleLocator(1))
if 0 < max_year-min_year < 10:
ax.xaxis.set_major_locator(MultipleLocator(1))
ax.xaxis.set_minor_locator(MultipleLocator(1))
# End for (case loop)
# End for (variables loop)
# Set up legend
# Gather labels based on case names and plotted line format (color, style, etc)
lines_labels = [ax.get_legend_handles_labels() for ax in fig.axes]
lines, labels = [sum(lol, []) for lol in zip(*lines_labels)]
fig.legend(lines[:case_names_len+1], labels[:case_names_len+1],
loc="center left",fontsize=18,
bbox_to_anchor=(0.365, 0.4,.02,.05)) #bbox_to_anchor(x0, y0, width, height)
fig.show()
#plt.savefig("TimeSeries_ANN.png", facecolor='w',bbox_inches="tight")
Plotting variable: RESTOM
Plotting variable: TS
Plotting variable: ICEFRAC
FigureCanvasAgg is non-interactive, and thus cannot be shown