Analysis of Surface Fields#
mom6_tools.MOM6grid returns an object with MOM6 grid data.
mom6_tools.latlon_analysis has a collection of tools used to perform spatial analysis (e.g., time averages and spatial mean).
The goal of this notebook is the following:
server as an example of how to post-process CESM/MOM6 output;
create time averages of surface fields;
create time-series of globally-averaged surface fields;
%load_ext autoreload
%autoreload 2
import xarray as xr
import warnings, os
import intake
from datetime import datetime
from mom6_tools.MOM6grid import MOM6grid
from mom6_tools.surface import get_MLD, get_BLD
warnings.filterwarnings("ignore")
Basemap module not found. Some regional plots may not function properly
CESM_output_dir = "/glade/campaign/cesm/development/cross-wg/diagnostic_framework/CESM_output_for_testing"
Case = "b.e23_alpha16b.BLT1850.ne30_t232.054"
savefigs = False
mom6_tools_config = {}
# Parameters
CESM_output_dir = "/glade/campaign/cesm/development/cross-wg/diagnostic_framework/CESM_output_for_testing"
Case = "b.e23_alpha16b.BLT1850.ne30_t232.054"
savefigs = False
mom6_tools_config = {
"start_date": "0091-01-01",
"end_date": "0101-01-01",
"Fnames": {"native": "mom6.h.native.????-??.nc", "static": "mom6.h.static.nc"},
"oce_cat": "/glade/u/home/gmarques/libs/oce-catalogs/reference-datasets.yml",
}
subset_kwargs = {}
product = "/glade/u/home/dbailey/CUPiD/examples/coupled_model/computed_notebooks/quick-run/ocean_surface.ipynb"
OUTDIR = f'{CESM_output_dir}/{Case}/ocn/hist/'
print('Output directory is:', OUTDIR)
Output directory is: /glade/campaign/cesm/development/cross-wg/diagnostic_framework/CESM_output_for_testing/b.e23_alpha16b.BLT1850.ne30_t232.054/ocn/hist/
# The following parameters must be set accordingly
######################################################
# create an empty class object
class args:
pass
args.start_date = mom6_tools_config['start_date']
args.end_date = mom6_tools_config['end_date']
args.casename = Case
args.native = f"{Case}.{mom6_tools_config['Fnames']['native']}"
args.static = f"{Case}.{mom6_tools_config['Fnames']['static']}"
args.mld_obs = "mld-deboyer-tx2_3v2"
args.savefigs = savefigs
if not os.path.isdir('PNG/BLD'):
print('Creating a directory to place figures (PNG/BLD)... \n')
os.system('mkdir -p PNG/BLD')
if not os.path.isdir('PNG/MLD'):
print('Creating a directory to place figures (PNG/MLD)... \n')
os.system('mkdir -p PNG/MLD')
if not os.path.isdir('ncfiles'):
print('Creating a directory to place netcdf files (ncfiles)... \n')
os.system('mkdir ncfiles')
Creating a directory to place figures (PNG/BLD)...
Creating a directory to place figures (PNG/MLD)...
Creating a directory to place netcdf files (ncfiles)...
# load mom6 grid
grd = MOM6grid(OUTDIR+args.static)
grd_xr = MOM6grid(OUTDIR+args.static, xrformat=True)
MOM6 grid successfully loaded...
MOM6 grid successfully loaded...
print('Reading native dataset...')
startTime = datetime.now()
def preprocess(ds):
''' Compute montly averages and return the dataset with variables'''
variables = ['oml','mlotst','tos','SSH', 'SSU', 'SSV', 'speed', 'time_bnds']
for v in variables:
if v not in ds.variables:
ds[v] = xr.zeros_like(ds.SSH)
return ds[variables]
ds1 = xr.open_mfdataset(OUTDIR+args.native, parallel=False)
ds = preprocess(ds1)
print('Time elasped: ', datetime.now() - startTime)
Reading native dataset...
Time elasped: 0:03:40.243508
print('Selecting data between {} and {}...'.format(args.start_date, args.end_date))
ds_sel = ds.sel(time=slice(args.start_date, args.end_date))
Selecting data between 0091-01-01 and 0101-01-01...
catalog = intake.open_catalog(mom6_tools_config['oce_cat'])
mld_obs = catalog[args.mld_obs].to_dask()
# uncomment to list all datasets available
#list(catalog)
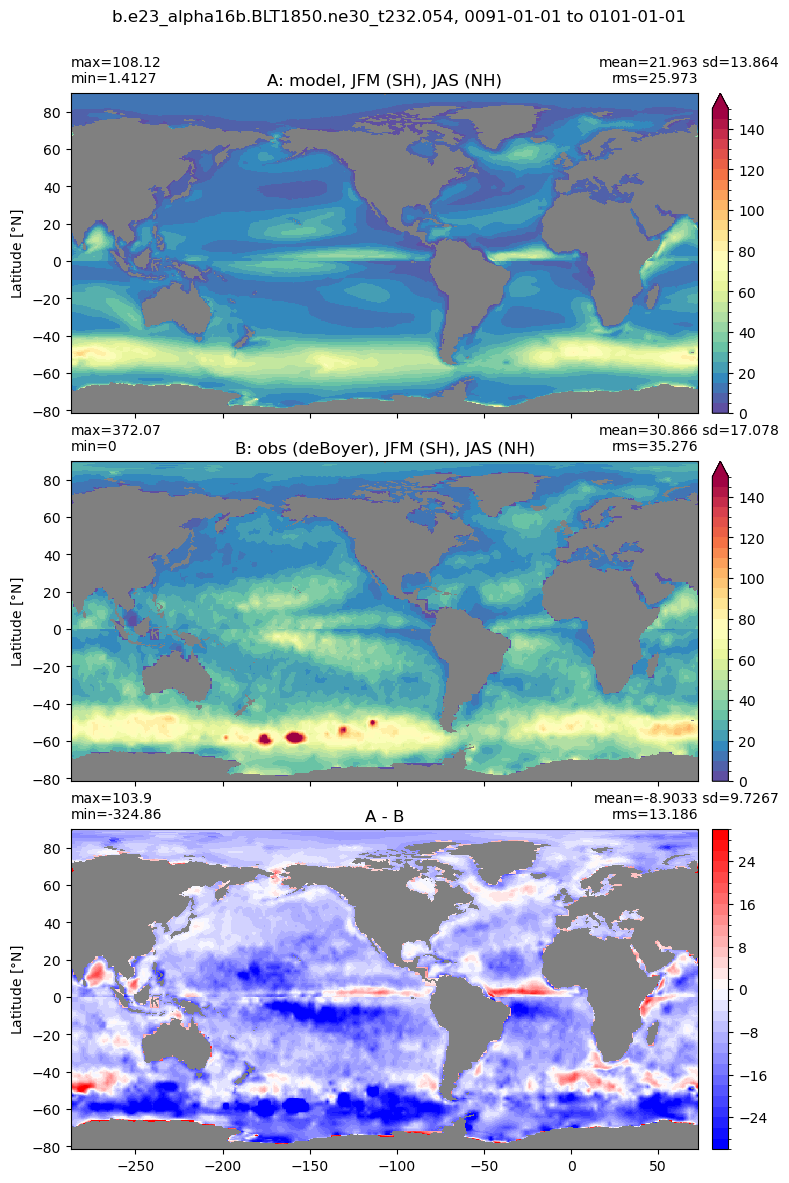
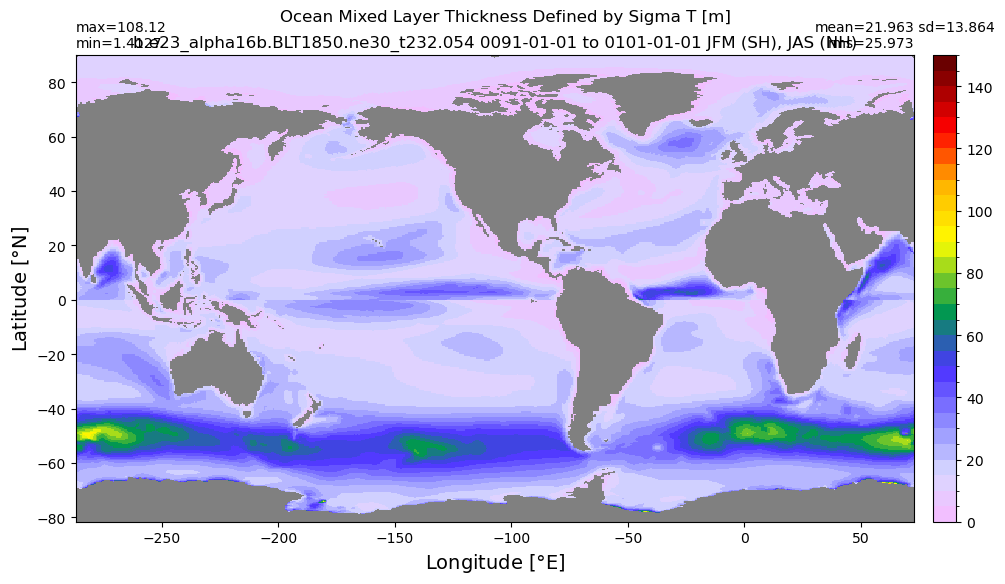
Mixed layer depth#
%matplotlib inline
# MLD
get_MLD(ds,'mlotst', mld_obs, grd, args)
Computing monthly MLD climatology...
Time elasped: 0:02:20.530319
Plotting...
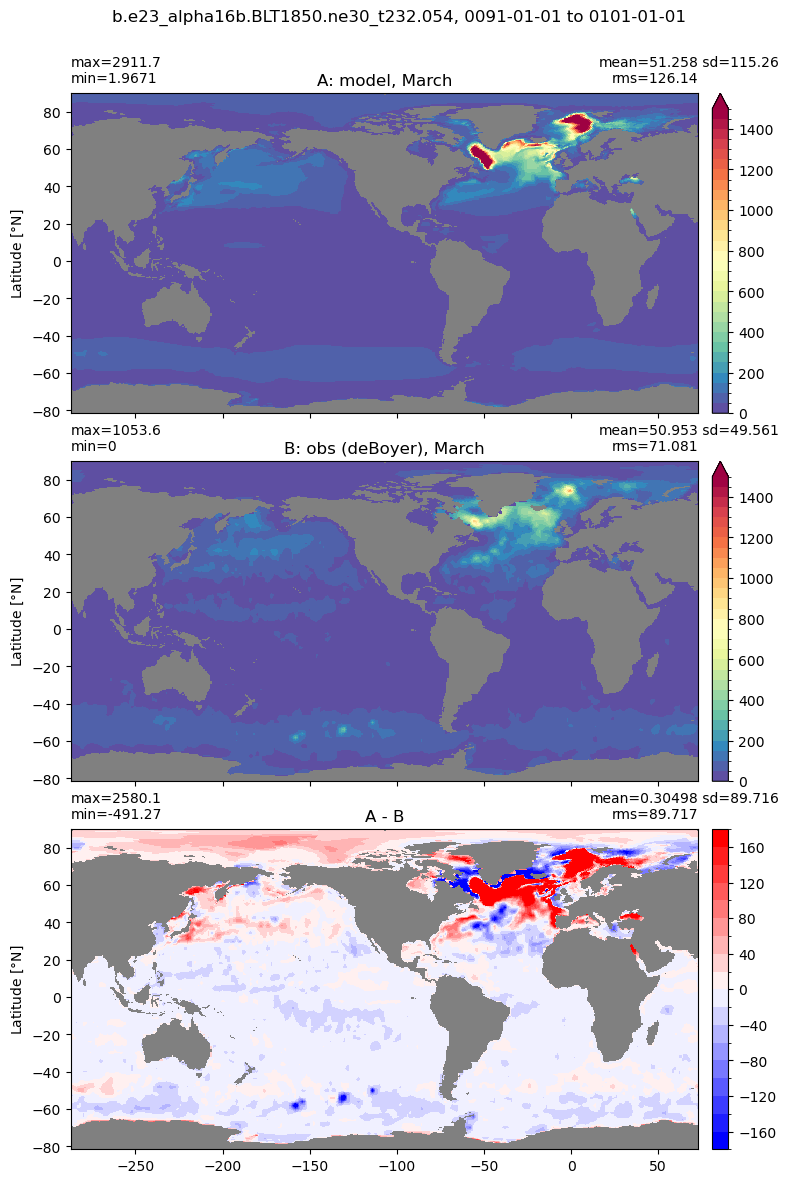
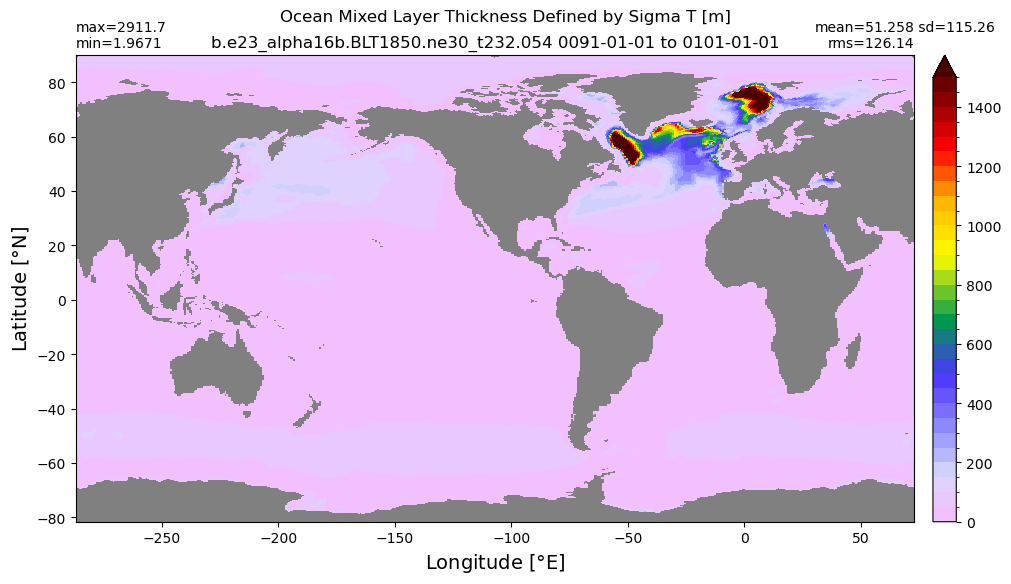
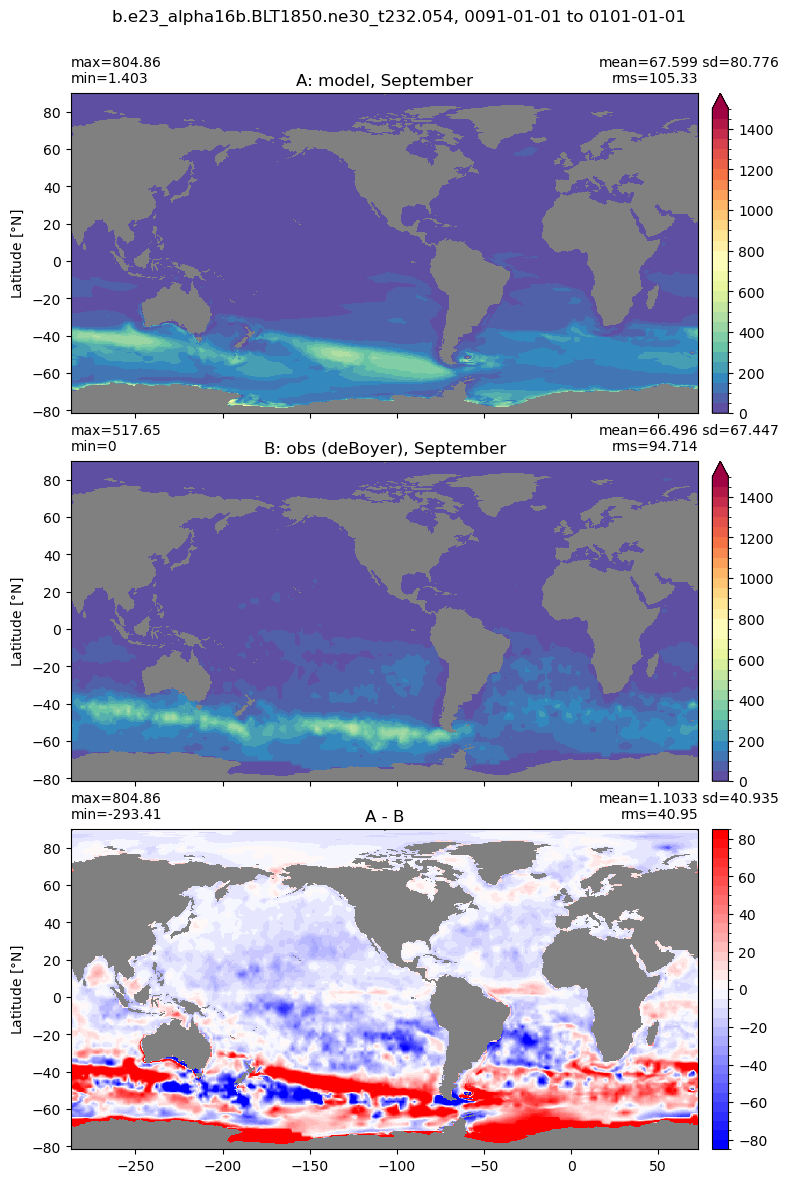
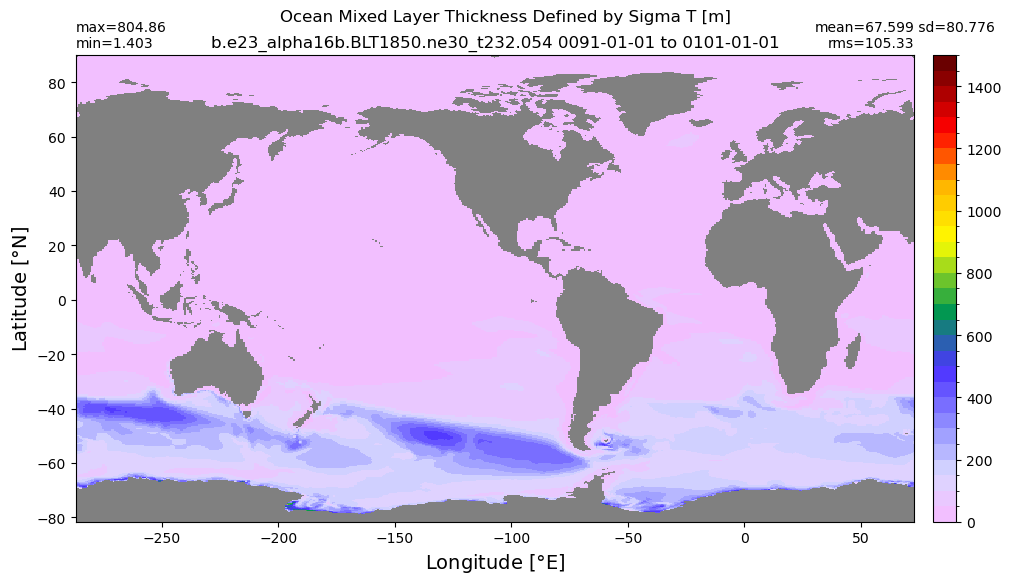
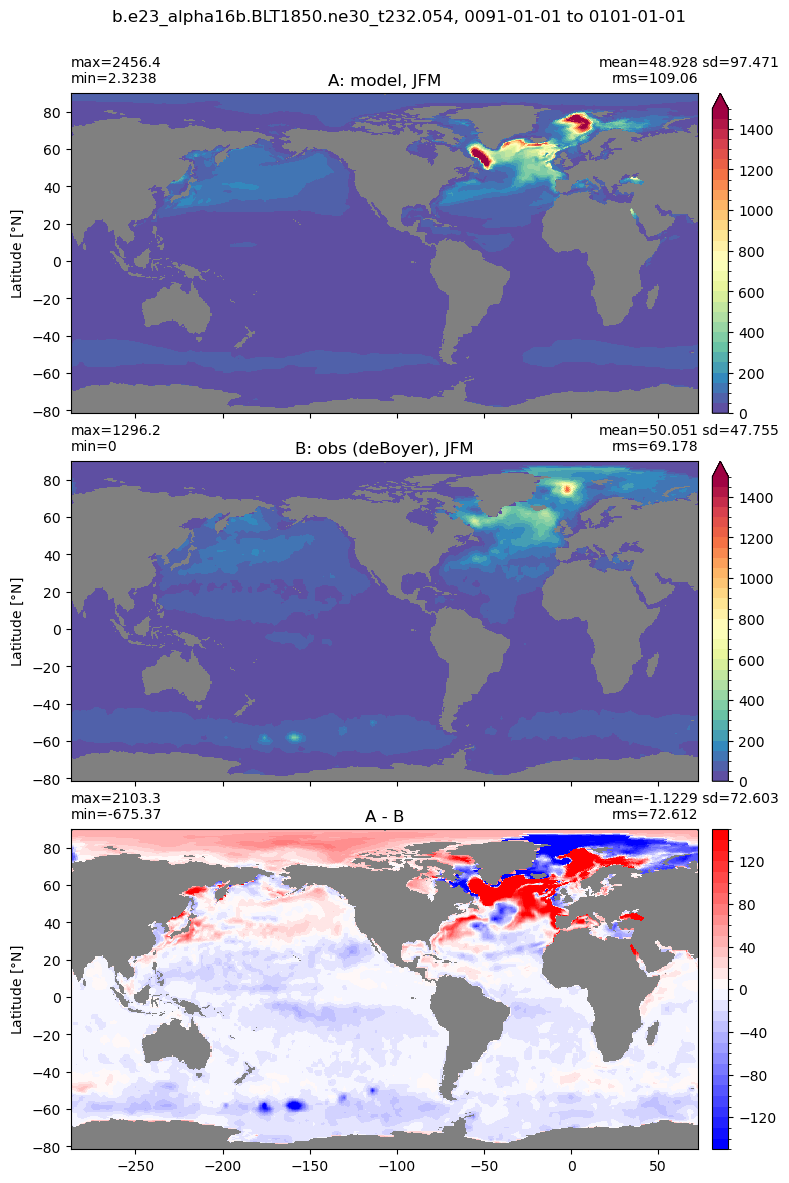
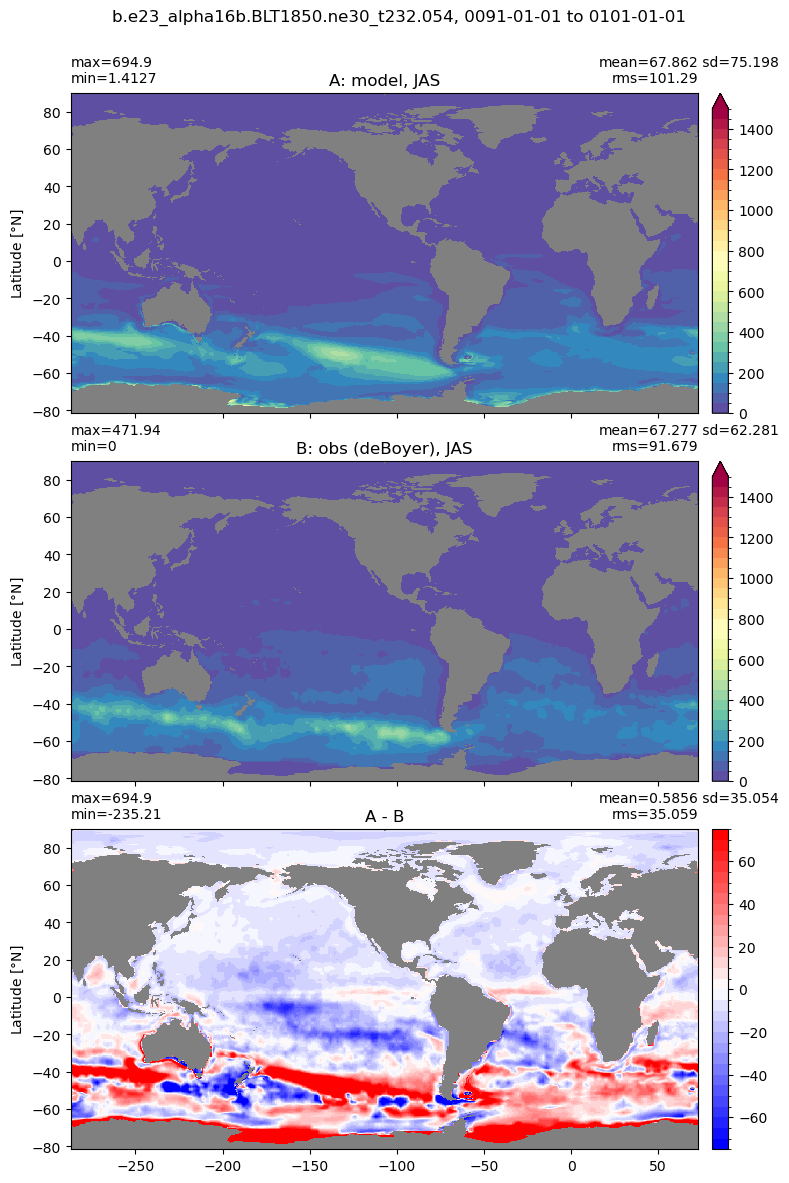
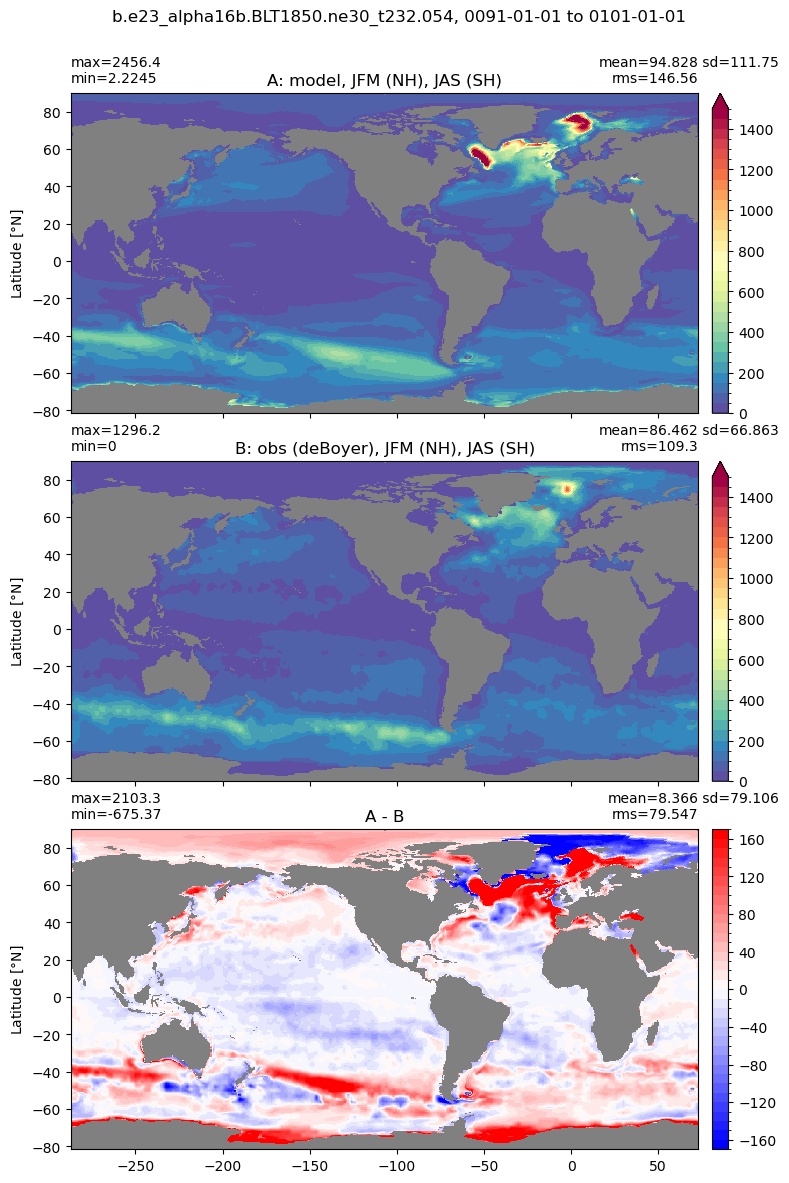
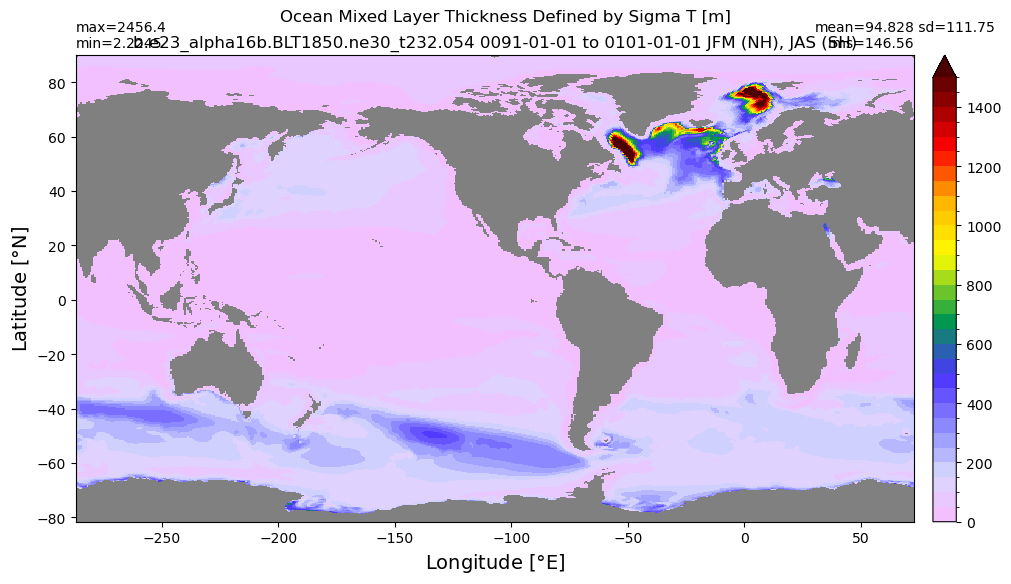
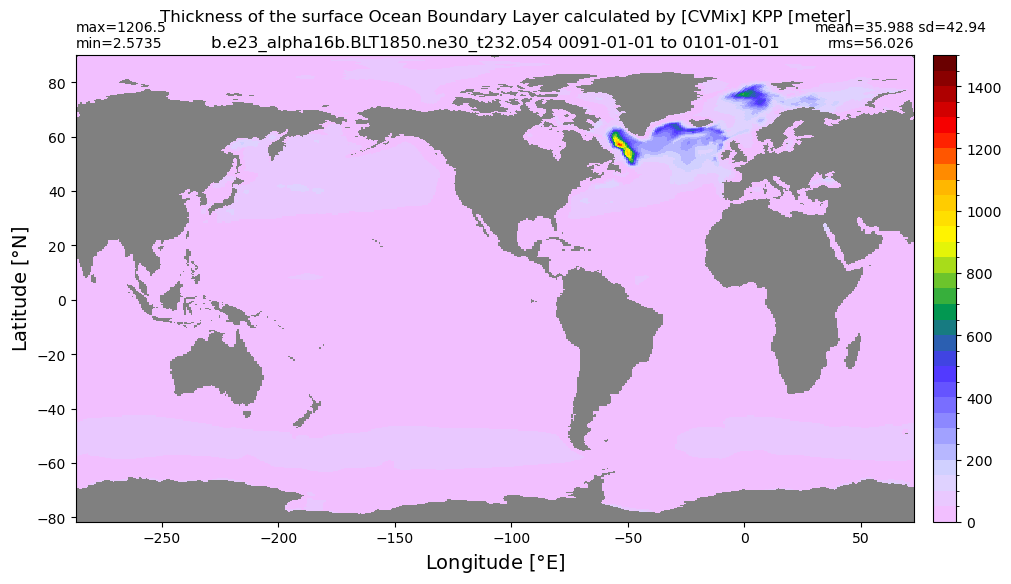
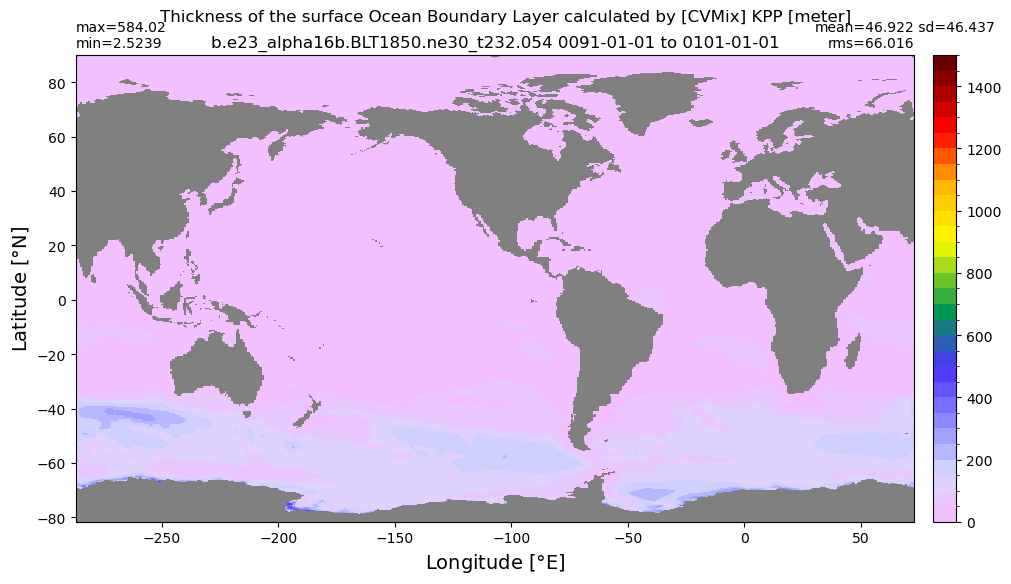
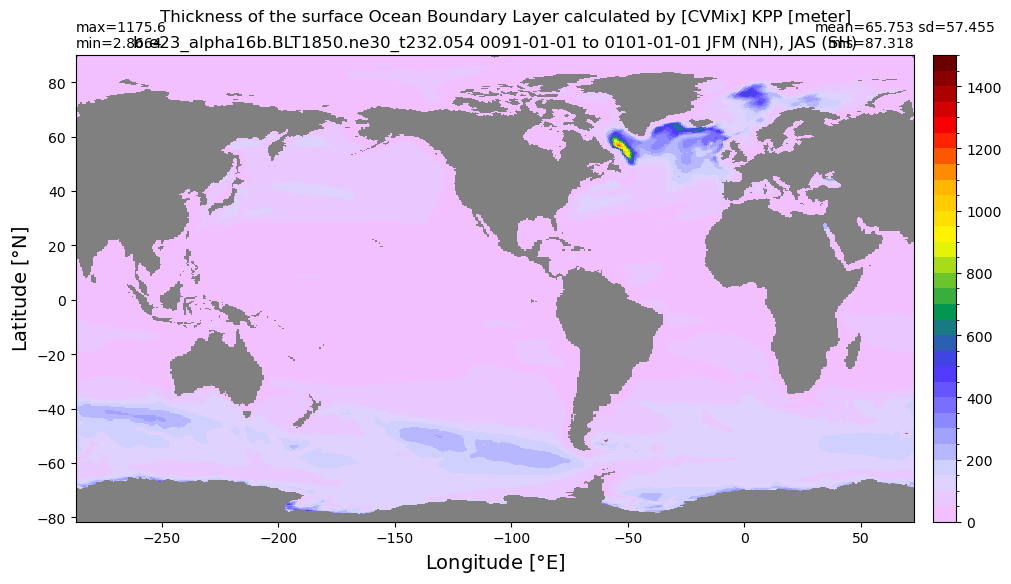
Boundary layer depth#
get_BLD(ds, 'oml', grd, args)
Computing monthly BLD climatology...
Time elasped: 0:02:23.782546
Plotting...
# SSH (not working)
#get_SSH(ds, 'SSH', grd, args)
