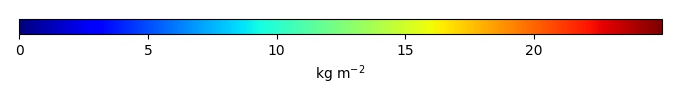
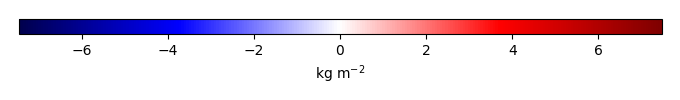
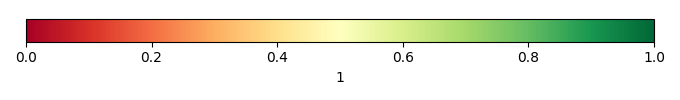Mean State
Download Data |
Period Mean (original grids) [Pg] |
Model Period Mean (intersection) [Pg] |
Model Period Mean (complement) [Pg] |
Benchmark Period Mean (intersection) [Pg] |
Benchmark Period Mean (complement) [Pg] |
Bias [kg m-2] |
Bias Score [1] |
Spatial Distribution Score [1] |
Overall Score [1] |
|||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Benchmark | [-] | 6.34 | ||||||||||
| CRUNCEPv7 | [-] | 8.25 | 7.39 | 0.731 | 6.34 | 0.191 | 0.499 | 0.59 | 0.83 | 0.71 | ||
| GSWP3v1 | [-] | 8.00 | 7.13 | 0.773 | 6.34 | 0.191 | 0.497 | 0.60 | 0.84 | 0.72 | ||
| WATCH | [-] | 9.03 | 8.18 | 0.749 | 6.34 | 0.191 | 0.637 | 0.61 | 0.84 | 0.72 |
Download Data |
Period Mean (original grids) [Pg] |
Model Period Mean (intersection) [Pg] |
Model Period Mean (complement) [Pg] |
Benchmark Period Mean (intersection) [Pg] |
Benchmark Period Mean (complement) [Pg] |
Bias [kg m-2] |
Bias Score [1] |
Spatial Distribution Score [1] |
Overall Score [1] |
|||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Benchmark | [-] | 36.8 | ||||||||||
| CRUNCEPv7 | [-] | 68.0 | 67.8 | 0.253 | 37.9 | 0.0273 | 2.60 | 0.49 | 0.52 | 0.50 | ||
| GSWP3v1 | [-] | 48.6 | 48.3 | 0.231 | 37.9 | 0.0273 | 0.941 | 0.47 | 0.65 | 0.56 | ||
| WATCH | [-] | 44.8 | 44.5 | 0.177 | 37.9 | 0.0273 | 0.606 | 0.66 | 0.92 | 0.79 |
Download Data |
Period Mean (original grids) [Pg] |
Model Period Mean (intersection) [Pg] |
Model Period Mean (complement) [Pg] |
Benchmark Period Mean (intersection) [Pg] |
Benchmark Period Mean (complement) [Pg] |
Bias [kg m-2] |
Bias Score [1] |
Spatial Distribution Score [1] |
Overall Score [1] |
|||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Benchmark | [-] | 19.1 | ||||||||||
| CRUNCEPv7 | [-] | 54.2 | 54.1 | 0.371 | 19.0 | 0.240 | 3.94 | 0.31 | 0.48 | 0.39 | ||
| GSWP3v1 | [-] | 45.7 | 45.6 | 0.298 | 19.0 | 0.240 | 2.99 | 0.37 | 0.56 | 0.47 | ||
| WATCH | [-] | 39.3 | 39.2 | 0.280 | 19.0 | 0.240 | 2.28 | 0.45 | 0.66 | 0.55 |
Download Data |
Period Mean (original grids) [Pg] |
Model Period Mean (intersection) [Pg] |
Model Period Mean (complement) [Pg] |
Benchmark Period Mean (intersection) [Pg] |
Benchmark Period Mean (complement) [Pg] |
Bias [kg m-2] |
Bias Score [1] |
Spatial Distribution Score [1] |
Overall Score [1] |
|||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Benchmark | [-] | 7.57 | ||||||||||
| CRUNCEPv7 | [-] | 10.2 | 9.47 | 0.496 | 7.28 | 0.0545 | 1.74 | 0.53 | 0.56 | 0.54 | ||
| GSWP3v1 | [-] | 8.57 | 7.92 | 0.448 | 7.28 | 0.0545 | 1.07 | 0.56 | 0.67 | 0.62 | ||
| WATCH | [-] | 9.34 | 8.78 | 0.430 | 7.28 | 0.0545 | 1.41 | 0.56 | 0.66 | 0.61 |
Download Data |
Period Mean (original grids) [Pg] |
Model Period Mean (intersection) [Pg] |
Model Period Mean (complement) [Pg] |
Benchmark Period Mean (intersection) [Pg] |
Benchmark Period Mean (complement) [Pg] |
Bias [kg m-2] |
Bias Score [1] |
Spatial Distribution Score [1] |
Overall Score [1] |
|||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Benchmark | [-] | 34.1 | ||||||||||
| CRUNCEPv7 | [-] | 37.8 | 36.3 | 1.08 | 33.6 | 0.103 | 0.429 | 0.54 | 0.78 | 0.66 | ||
| GSWP3v1 | [-] | 32.2 | 30.9 | 0.916 | 33.6 | 0.103 | 0.0802 | 0.58 | 0.85 | 0.71 | ||
| WATCH | [-] | 36.5 | 35.2 | 0.963 | 33.6 | 0.103 | 0.300 | 0.64 | 0.90 | 0.77 |
Download Data |
Period Mean (original grids) [Pg] |
Model Period Mean (intersection) [Pg] |
Model Period Mean (complement) [Pg] |
Benchmark Period Mean (intersection) [Pg] |
Benchmark Period Mean (complement) [Pg] |
Bias [kg m-2] |
Bias Score [1] |
Spatial Distribution Score [1] |
Overall Score [1] |
|||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Benchmark | [-] | 28.1 | ||||||||||
| CRUNCEPv7 | [-] | 49.8 | 46.9 | 2.23 | 26.7 | 1.53 | 11.2 | 0.46 | 0.90 | 0.68 | ||
| GSWP3v1 | [-] | 40.3 | 38.0 | 1.80 | 26.7 | 1.53 | 7.81 | 0.60 | 0.92 | 0.76 | ||
| WATCH | [-] | 39.2 | 36.9 | 1.83 | 26.7 | 1.53 | 7.53 | 0.63 | 0.88 | 0.76 |
Download Data |
Period Mean (original grids) [Pg] |
Model Period Mean (intersection) [Pg] |
Model Period Mean (complement) [Pg] |
Benchmark Period Mean (intersection) [Pg] |
Benchmark Period Mean (complement) [Pg] |
Bias [kg m-2] |
Bias Score [1] |
Spatial Distribution Score [1] |
Overall Score [1] |
|||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Benchmark | [-] | 13.6 | ||||||||||
| CRUNCEPv7 | [-] | 30.4 | 29.4 | 0.754 | 13.5 | 0.0773 | 3.29 | 0.43 | 0.26 | 0.35 | ||
| GSWP3v1 | [-] | 20.7 | 19.9 | 0.628 | 13.5 | 0.0773 | 1.72 | 0.47 | 0.35 | 0.41 | ||
| WATCH | [-] | 15.4 | 14.7 | 0.552 | 13.5 | 0.0773 | 0.883 | 0.52 | 0.53 | 0.53 |
Download Data |
Period Mean (original grids) [Pg] |
Model Period Mean (intersection) [Pg] |
Model Period Mean (complement) [Pg] |
Benchmark Period Mean (intersection) [Pg] |
Benchmark Period Mean (complement) [Pg] |
Bias [kg m-2] |
Bias Score [1] |
Spatial Distribution Score [1] |
Overall Score [1] |
|||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Benchmark | [-] | 368. | ||||||||||
| CRUNCEPv7 | [-] | 589. | 578. | 11.2 | 363. | 4.64 | 2.37 | 0.51 | 0.68 | 0.60 | ||
| GSWP3v1 | [-] | 487. | 477. | 9.77 | 363. | 4.64 | 1.46 | 0.57 | 0.78 | 0.68 | ||
| WATCH | [-] | 505. | 496. | 9.63 | 363. | 4.64 | 1.60 | 0.60 | 0.81 | 0.70 |
Download Data |
Period Mean (original grids) [Pg] |
Model Period Mean (intersection) [Pg] |
Model Period Mean (complement) [Pg] |
Benchmark Period Mean (intersection) [Pg] |
Benchmark Period Mean (complement) [Pg] |
Bias [kg m-2] |
Bias Score [1] |
Spatial Distribution Score [1] |
Overall Score [1] |
|||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Benchmark | [-] | 1.17 | ||||||||||
| CRUNCEPv7 | [-] | 1.86 | 1.51 | 0.341 | 1.14 | 0.0276 | 0.219 | 0.50 | 0.65 | 0.58 | ||
| GSWP3v1 | [-] | 1.56 | 1.27 | 0.276 | 1.14 | 0.0276 | 0.173 | 0.50 | 0.63 | 0.56 | ||
| WATCH | [-] | 2.12 | 1.76 | 0.337 | 1.14 | 0.0276 | 0.302 | 0.50 | 0.55 | 0.52 |
Download Data |
Period Mean (original grids) [Pg] |
Model Period Mean (intersection) [Pg] |
Model Period Mean (complement) [Pg] |
Benchmark Period Mean (intersection) [Pg] |
Benchmark Period Mean (complement) [Pg] |
Bias [kg m-2] |
Bias Score [1] |
Spatial Distribution Score [1] |
Overall Score [1] |
|||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Benchmark | [-] | 26.7 | ||||||||||
| CRUNCEPv7 | [-] | 29.2 | 26.1 | 0.472 | 26.6 | 0.0438 | 0.0763 | 0.60 | 0.84 | 0.72 | ||
| GSWP3v1 | [-] | 27.1 | 24.2 | 0.487 | 26.6 | 0.0438 | -0.137 | 0.64 | 0.89 | 0.76 | ||
| WATCH | [-] | 30.1 | 27.2 | 0.509 | 26.6 | 0.0438 | 0.186 | 0.67 | 0.90 | 0.79 |
Download Data |
Period Mean (original grids) [Pg] |
Model Period Mean (intersection) [Pg] |
Model Period Mean (complement) [Pg] |
Benchmark Period Mean (intersection) [Pg] |
Benchmark Period Mean (complement) [Pg] |
Bias [kg m-2] |
Bias Score [1] |
Spatial Distribution Score [1] |
Overall Score [1] |
|||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Benchmark | [-] | 28.3 | ||||||||||
| CRUNCEPv7 | [-] | 51.3 | 46.5 | 0.541 | 28.2 | 0.0700 | 7.29 | 0.58 | 0.90 | 0.74 | ||
| GSWP3v1 | [-] | 44.0 | 40.1 | 0.477 | 28.2 | 0.0700 | 5.08 | 0.64 | 0.93 | 0.79 | ||
| WATCH | [-] | 46.6 | 42.1 | 0.449 | 28.2 | 0.0700 | 5.70 | 0.63 | 0.93 | 0.78 |
Download Data |
Period Mean (original grids) [Pg] |
Model Period Mean (intersection) [Pg] |
Model Period Mean (complement) [Pg] |
Benchmark Period Mean (intersection) [Pg] |
Benchmark Period Mean (complement) [Pg] |
Bias [kg m-2] |
Bias Score [1] |
Spatial Distribution Score [1] |
Overall Score [1] |
|||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Benchmark | [-] | 31.5 | ||||||||||
| CRUNCEPv7 | [-] | 35.1 | 34.5 | 0.777 | 30.5 | 0.349 | 1.20 | 0.53 | 0.79 | 0.66 | ||
| GSWP3v1 | [-] | 33.4 | 32.9 | 0.692 | 30.5 | 0.349 | 0.914 | 0.64 | 0.88 | 0.76 | ||
| WATCH | [-] | 33.4 | 32.9 | 0.703 | 30.5 | 0.349 | 0.917 | 0.64 | 0.89 | 0.77 |
Download Data |
Period Mean (original grids) [Pg] |
Model Period Mean (intersection) [Pg] |
Model Period Mean (complement) [Pg] |
Benchmark Period Mean (intersection) [Pg] |
Benchmark Period Mean (complement) [Pg] |
Bias [kg m-2] |
Bias Score [1] |
Spatial Distribution Score [1] |
Overall Score [1] |
|||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Benchmark | [-] | 23.7 | ||||||||||
| CRUNCEPv7 | [-] | 31.5 | 33.8 | 0.295 | 23.7 | 0.0475 | 1.12 | 0.60 | 0.84 | 0.72 | ||
| GSWP3v1 | [-] | 24.7 | 26.8 | 0.291 | 23.7 | 0.0475 | 0.408 | 0.64 | 0.90 | 0.77 | ||
| WATCH | [-] | 33.2 | 35.3 | 0.303 | 23.7 | 0.0475 | 1.30 | 0.63 | 0.89 | 0.76 |
Download Data |
Period Mean (original grids) [Pg] |
Model Period Mean (intersection) [Pg] |
Model Period Mean (complement) [Pg] |
Benchmark Period Mean (intersection) [Pg] |
Benchmark Period Mean (complement) [Pg] |
Bias [kg m-2] |
Bias Score [1] |
Spatial Distribution Score [1] |
Overall Score [1] |
|||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Benchmark | [-] | 72.3 | ||||||||||
| CRUNCEPv7 | [-] | 128. | 132. | 0.623 | 72.2 | 0.0879 | 4.19 | 0.51 | 0.78 | 0.64 | ||
| GSWP3v1 | [-] | 113. | 116. | 0.571 | 72.2 | 0.0879 | 3.13 | 0.60 | 0.87 | 0.73 | ||
| WATCH | [-] | 128. | 131. | 0.525 | 72.2 | 0.0879 | 4.09 | 0.52 | 0.81 | 0.66 |
Download Data |
Period Mean (original grids) [Pg] |
Model Period Mean (intersection) [Pg] |
Model Period Mean (complement) [Pg] |
Benchmark Period Mean (intersection) [Pg] |
Benchmark Period Mean (complement) [Pg] |
Bias [kg m-2] |
Bias Score [1] |
Spatial Distribution Score [1] |
Overall Score [1] |
|||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Benchmark | [-] | 27.3 | ||||||||||
| CRUNCEPv7 | [-] | 35.4 | 34.7 | 0.307 | 27.4 | 0.0588 | 0.996 | 0.54 | 0.73 | 0.64 | ||
| GSWP3v1 | [-] | 24.4 | 23.8 | 0.251 | 27.4 | 0.0588 | -0.212 | 0.58 | 0.85 | 0.71 | ||
| WATCH | [-] | 25.1 | 24.6 | 0.238 | 27.4 | 0.0588 | -0.134 | 0.62 | 0.86 | 0.74 |
Temporally integrated period mean