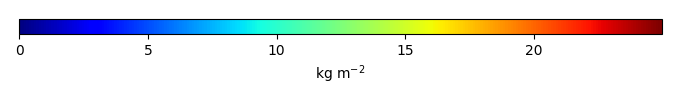
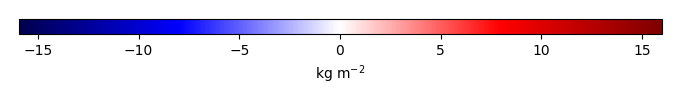
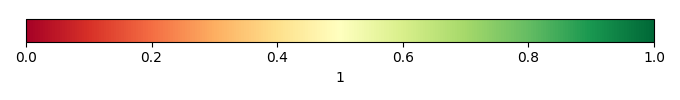Mean State
Download Data |
Period Mean (original grids) [Pg] |
Model Period Mean (intersection) [Pg] |
Model Period Mean (complement) [Pg] |
Benchmark Period Mean (intersection) [Pg] |
Benchmark Period Mean (complement) [Pg] |
Bias [kg m-2] |
Bias Score [1] |
Spatial Distribution Score [1] |
Overall Score [1] |
|||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Benchmark | [-] | 2.76 | ||||||||||
| CRUNCEPv7 | [-] | 54.2 | 7.41 | 47.0 | 2.66 | 0.105 | 4.50 | 0.42 | 0.73 | 0.58 | ||
| GSWP3v1 | [-] | 45.7 | 7.17 | 38.7 | 2.66 | 0.105 | 4.09 | 0.43 | 0.84 | 0.63 | ||
| WATCH | [-] | 39.3 | 5.25 | 34.2 | 2.66 | 0.105 | 2.48 | 0.48 | 0.83 | 0.66 |
Download Data |
Period Mean (original grids) [Pg] |
Model Period Mean (intersection) [Pg] |
Model Period Mean (complement) [Pg] |
Benchmark Period Mean (intersection) [Pg] |
Benchmark Period Mean (complement) [Pg] |
Bias [kg m-2] |
Bias Score [1] |
Spatial Distribution Score [1] |
Overall Score [1] |
|||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Benchmark | [-] | 0.878 | ||||||||||
| CRUNCEPv7 | [-] | 10.2 | 0.761 | 9.21 | 0.588 | 0.00736 | 1.48 | 0.63 | 0.84 | 0.74 | ||
| GSWP3v1 | [-] | 8.57 | 0.454 | 7.92 | 0.588 | 0.00736 | 0.419 | 0.59 | 0.83 | 0.71 | ||
| WATCH | [-] | 9.34 | 0.604 | 8.60 | 0.588 | 0.00736 | 0.846 | 0.63 | 0.88 | 0.75 |
Download Data |
Period Mean (original grids) [Pg] |
Model Period Mean (intersection) [Pg] |
Model Period Mean (complement) [Pg] |
Benchmark Period Mean (intersection) [Pg] |
Benchmark Period Mean (complement) [Pg] |
Bias [kg m-2] |
Bias Score [1] |
Spatial Distribution Score [1] |
Overall Score [1] |
|||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Benchmark | [-] | 30.1 | ||||||||||
| CRUNCEPv7 | [-] | 589. | 29.0 | 560. | 29.9 | 0.195 | 0.319 | 0.60 | 0.84 | 0.72 | ||
| GSWP3v1 | [-] | 487. | 22.3 | 465. | 29.9 | 0.195 | -0.481 | 0.58 | 0.77 | 0.67 | ||
| WATCH | [-] | 505. | 22.6 | 483. | 29.9 | 0.195 | -0.504 | 0.61 | 0.74 | 0.67 |
Download Data |
Period Mean (original grids) [Pg] |
Model Period Mean (intersection) [Pg] |
Model Period Mean (complement) [Pg] |
Benchmark Period Mean (intersection) [Pg] |
Benchmark Period Mean (complement) [Pg] |
Bias [kg m-2] |
Bias Score [1] |
Spatial Distribution Score [1] |
Overall Score [1] |
|||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Benchmark | [-] | 26.5 | ||||||||||
| CRUNCEPv7 | [-] | 35.4 | 20.9 | 14.1 | 26.7 | 0.0803 | -0.528 | 0.61 | 0.85 | 0.73 | ||
| GSWP3v1 | [-] | 24.4 | 14.7 | 9.36 | 26.7 | 0.0803 | -1.38 | 0.59 | 0.70 | 0.64 | ||
| WATCH | [-] | 25.1 | 16.7 | 8.18 | 26.7 | 0.0803 | -1.14 | 0.62 | 0.71 | 0.67 |
Temporally integrated period mean