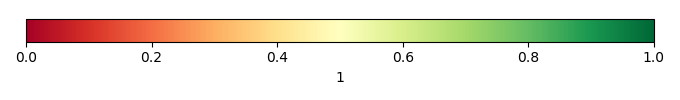Mean State
Download Data |
Period Mean (original grids) [Pg] |
Model Period Mean (intersection) [Pg] |
Model Period Mean (complement) [Pg] |
Benchmark Period Mean (intersection) [Pg] |
Benchmark Period Mean (complement) [Pg] |
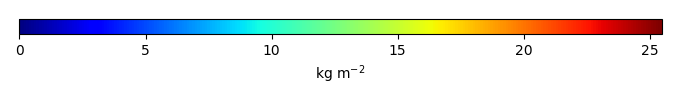
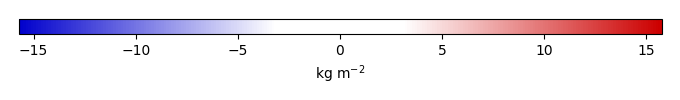
Bias [kg m-2] |
Bias Score [1] |
Spatial Distribution Score [1] |
Overall Score [1] |
|||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Benchmark | [-] | 26.2 | ||||||||||
| CLM5PHSOFF | [-] | 542. | 7.60 | 535. | 26.1 | 0.0878 | -2.26 | 0.46 | 0.50 | 0.48 | ||
| CLM5PHSON | [-] | 487. | 15.3 | 472. | 26.1 | 0.0878 | -1.07 | 0.60 | 0.79 | 0.69 |
Download Data |
Period Mean (original grids) [Pg] |
Model Period Mean (intersection) [Pg] |
Model Period Mean (complement) [Pg] |
Benchmark Period Mean (intersection) [Pg] |
Benchmark Period Mean (complement) [Pg] |
Bias [kg m-2] |
Bias Score [1] |
Spatial Distribution Score [1] |
Overall Score [1] |
|||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Benchmark | [-] | 0.00338 | ||||||||||
| CLM5PHSOFF | [-] | 16.3 | 0.0152 | 15.5 | 0.00338 | 0.00 | 0.116 | 0.58 | 0.77 | 0.67 | ||
| CLM5PHSON | [-] | 10.2 | 0.0260 | 9.91 | 0.00338 | 0.00 | 0.333 | 0.27 | 0.37 | 0.32 |
Download Data |
Period Mean (original grids) [Pg] |
Model Period Mean (intersection) [Pg] |
Model Period Mean (complement) [Pg] |
Benchmark Period Mean (intersection) [Pg] |
Benchmark Period Mean (complement) [Pg] |
Bias [kg m-2] |
Bias Score [1] |
Spatial Distribution Score [1] |
Overall Score [1] |
|||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Benchmark | [-] | 17.5 | ||||||||||
| CLM5PHSOFF | [-] | 7.11 | 5.93 | 0.828 | 17.4 | 0.0333 | -2.80 | 0.48 | 0.64 | 0.56 | ||
| CLM5PHSON | [-] | 12.0 | 11.6 | 0.234 | 17.4 | 0.0333 | -1.09 | 0.65 | 0.88 | 0.77 |
Download Data |
Period Mean (original grids) [Pg] |
Model Period Mean (intersection) [Pg] |
Model Period Mean (complement) [Pg] |
Benchmark Period Mean (intersection) [Pg] |
Benchmark Period Mean (complement) [Pg] |
Bias [kg m-2] |
Bias Score [1] |
Spatial Distribution Score [1] |
Overall Score [1] |
|||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Benchmark | [-] | 0.222 | ||||||||||
| CLM5PHSOFF | [-] | 32.6 | 0.287 | 32.3 | 0.222 | 0.00 | 0.504 | 0.49 | 0.64 | 0.56 | ||
| CLM5PHSON | [-] | 43.6 | 0.598 | 43.2 | 0.222 | 0.00 | 4.61 | 0.41 | 0.64 | 0.53 |
Download Data |
Period Mean (original grids) [Pg] |
Model Period Mean (intersection) [Pg] |
Model Period Mean (complement) [Pg] |
Benchmark Period Mean (intersection) [Pg] |
Benchmark Period Mean (complement) [Pg] |
Bias [kg m-2] |
Bias Score [1] |
Spatial Distribution Score [1] |
Overall Score [1] |
|||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Benchmark | [-] | 8.47 | ||||||||||
| CLM5PHSOFF | [-] | 1.45 | 1.37 | 0.175 | 8.42 | 0.0520 | -1.80 | 0.41 | 0.29 | 0.35 | ||
| CLM5PHSON | [-] | 3.04 | 3.04 | 0.0355 | 8.42 | 0.0520 | -1.23 | 0.50 | 0.55 | 0.52 |
Temporally integrated period mean