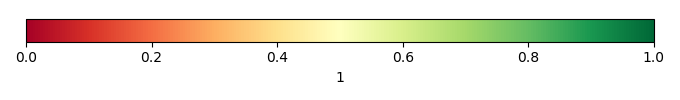Mean State
Download Data |
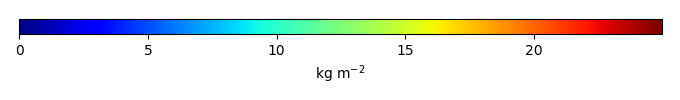
Period Mean (original grids) [Pg] |
Model Period Mean (intersection) [Pg] |
Model Period Mean (complement) [Pg] |
Benchmark Period Mean (intersection) [Pg] |
Benchmark Period Mean (complement) [Pg] |
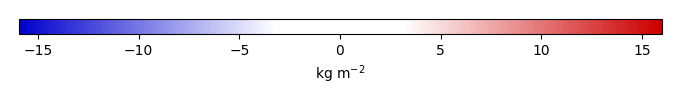
Bias [kg m-2] |
Bias Score [1] |
Spatial Distribution Score [1] |
Overall Score [1] |
|||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Benchmark | [-] | 30.1 | ||||||||||
| CRUNCEPv7 | [-] | 589. | 29.0 | 560. | 29.9 | 0.195 | 0.319 | 0.60 | 0.84 | 0.72 | ||
| GSWP3v1 | [-] | 487. | 22.3 | 465. | 29.9 | 0.195 | -0.481 | 0.58 | 0.77 | 0.67 | ||
| WATCH | [-] | 505. | 22.6 | 483. | 29.9 | 0.195 | -0.504 | 0.61 | 0.74 | 0.67 |
Download Data |
Period Mean (original grids) [Pg] |
Model Period Mean (intersection) [Pg] |
Model Period Mean (complement) [Pg] |
Benchmark Period Mean (intersection) [Pg] |
Benchmark Period Mean (complement) [Pg] |
Bias [kg m-2] |
Bias Score [1] |
Spatial Distribution Score [1] |
Overall Score [1] |
|||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Benchmark | [-] | 1.44 | ||||||||||
| CRUNCEPv7 | [-] | 16.4 | 5.84 | 10.4 | 1.41 | 0.0279 | 4.22 | 0.26 | 0.35 | 0.31 | ||
| GSWP3v1 | [-] | 14.5 | 5.95 | 8.42 | 1.41 | 0.0279 | 4.18 | 0.21 | 0.47 | 0.34 | ||
| WATCH | [-] | 11.4 | 4.46 | 6.77 | 1.41 | 0.0279 | 2.85 | 0.31 | 0.55 | 0.43 |
Download Data |
Period Mean (original grids) [Pg] |
Model Period Mean (intersection) [Pg] |
Model Period Mean (complement) [Pg] |
Benchmark Period Mean (intersection) [Pg] |
Benchmark Period Mean (complement) [Pg] |
Bias [kg m-2] |
Bias Score [1] |
Spatial Distribution Score [1] |
Overall Score [1] |
|||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Benchmark | [-] | 0.00649 | ||||||||||
| CRUNCEPv7 | [-] | 11.9 | 0.0238 | 11.6 | 0.00649 | 0.00 | 0.192 | 0.54 | 0.67 | 0.61 | ||
| GSWP3v1 | [-] | 10.2 | 0.0250 | 9.91 | 0.00649 | 0.00 | 0.233 | 0.58 | 0.75 | 0.67 | ||
| WATCH | [-] | 10.8 | 0.0311 | 10.6 | 0.00649 | 0.00 | 0.336 | 0.78 | 0.78 | 0.78 |
Download Data |
Period Mean (original grids) [Pg] |
Model Period Mean (intersection) [Pg] |
Model Period Mean (complement) [Pg] |
Benchmark Period Mean (intersection) [Pg] |
Benchmark Period Mean (complement) [Pg] |
Bias [kg m-2] |
Bias Score [1] |
Spatial Distribution Score [1] |
Overall Score [1] |
|||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Benchmark | [-] | 17.4 | ||||||||||
| CRUNCEPv7 | [-] | 17.8 | 17.3 | 0.314 | 17.3 | 0.0180 | 0.419 | 0.69 | 0.92 | 0.80 | ||
| GSWP3v1 | [-] | 12.0 | 11.6 | 0.244 | 17.3 | 0.0180 | -1.06 | 0.65 | 0.85 | 0.75 | ||
| WATCH | [-] | 13.9 | 13.5 | 0.233 | 17.3 | 0.0180 | -0.616 | 0.70 | 0.86 | 0.78 |
Download Data |
Period Mean (original grids) [Pg] |
Model Period Mean (intersection) [Pg] |
Model Period Mean (complement) [Pg] |
Benchmark Period Mean (intersection) [Pg] |
Benchmark Period Mean (complement) [Pg] |
Bias [kg m-2] |
Bias Score [1] |
Spatial Distribution Score [1] |
Overall Score [1] |
|||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Benchmark | [-] | 1.52 | ||||||||||
| CRUNCEPv7 | [-] | 56.8 | 2.46 | 54.5 | 1.46 | 0.0621 | 7.01 | 0.56 | 0.88 | 0.72 | ||
| GSWP3v1 | [-] | 43.6 | 1.80 | 42.0 | 1.46 | 0.0621 | 3.72 | 0.65 | 0.90 | 0.77 | ||
| WATCH | [-] | 38.9 | 1.31 | 37.8 | 1.46 | 0.0621 | 1.14 | 0.64 | 0.74 | 0.69 |
Download Data |
Period Mean (original grids) [Pg] |
Model Period Mean (intersection) [Pg] |
Model Period Mean (complement) [Pg] |
Benchmark Period Mean (intersection) [Pg] |
Benchmark Period Mean (complement) [Pg] |
Bias [kg m-2] |
Bias Score [1] |
Spatial Distribution Score [1] |
Overall Score [1] |
|||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Benchmark | [-] | 9.78 | ||||||||||
| CRUNCEPv7 | [-] | 3.49 | 3.35 | 0.157 | 9.71 | 0.0683 | -1.61 | 0.49 | 0.57 | 0.53 | ||
| GSWP3v1 | [-] | 3.04 | 2.93 | 0.149 | 9.71 | 0.0683 | -1.73 | 0.48 | 0.41 | 0.45 | ||
| WATCH | [-] | 3.35 | 3.21 | 0.171 | 9.71 | 0.0683 | -1.67 | 0.48 | 0.43 | 0.45 |
Temporally integrated period mean