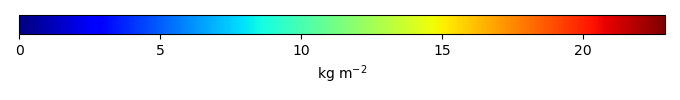
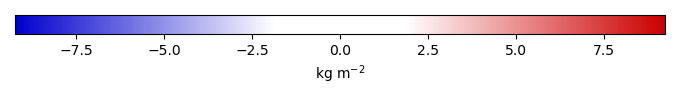
Mean State
Download Data |
Period Mean (original grids) [Pg] |
Model Period Mean (intersection) [Pg] |
Benchmark Period Mean (intersection) [Pg] |
Model Period Mean (complement) [Pg] |
Benchmark Period Mean (complement) [Pg] |
Bias [kg m-2] |
Bias Score [1] |
Spatial Distribution Score [1] |
Overall Score [1] |
|||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Benchmark | [-] | 26.2 | ||||||||||
| I123 | [-] | 463. | 22.7 | 26.2 | 438. | 0.000133 | -0.121 | 0.468 | 0.884 | 0.676 | ||
| post5.4 | [-] | 471. | 23.7 | 26.2 | 446. | 0.000133 | 0.0110 | 0.465 | 0.896 | 0.680 |
Download Data |
Period Mean (original grids) [Pg] |
Model Period Mean (intersection) [Pg] |
Benchmark Period Mean (intersection) [Pg] |
Model Period Mean (complement) [Pg] |
Benchmark Period Mean (complement) [Pg] |
Bias [kg m-2] |
Bias Score [1] |
Spatial Distribution Score [1] |
Overall Score [1] |
|||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Benchmark | [-] | 0.00338 | ||||||||||
| I123 | [-] | 10.9 | 0.0364 | 0.00338 | 10.5 | 0.512 | 0.0139 | 0.308 | 0.161 | |||
| post5.4 | [-] | 11.0 | 0.0373 | 0.00338 | 10.6 | 0.524 | 0.0142 | 0.310 | 0.162 |
Download Data |
Period Mean (original grids) [Pg] |
Model Period Mean (intersection) [Pg] |
Benchmark Period Mean (intersection) [Pg] |
Model Period Mean (complement) [Pg] |
Benchmark Period Mean (complement) [Pg] |
Bias [kg m-2] |
Bias Score [1] |
Spatial Distribution Score [1] |
Overall Score [1] |
|||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Benchmark | [-] | 17.5 | ||||||||||
| I123 | [-] | 16.8 | 16.3 | 17.5 | 0.322 | 6.03e-05 | 0.0890 | 0.614 | 0.925 | 0.770 | ||
| post5.4 | [-] | 17.5 | 17.1 | 17.5 | 0.335 | 6.03e-05 | 0.283 | 0.612 | 0.933 | 0.772 |
Download Data |
Period Mean (original grids) [Pg] |
Model Period Mean (intersection) [Pg] |
Benchmark Period Mean (intersection) [Pg] |
Model Period Mean (complement) [Pg] |
Benchmark Period Mean (complement) [Pg] |
Bias [kg m-2] |
Bias Score [1] |
Spatial Distribution Score [1] |
Overall Score [1] |
|||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Benchmark | [-] | 0.222 | ||||||||||
| I123 | [-] | 26.2 | 0.636 | 0.222 | 25.7 | 4.96 | 0.214 | 0.707 | 0.460 | |||
| post5.4 | [-] | 27.4 | 0.665 | 0.222 | 26.9 | 5.29 | 0.207 | 0.687 | 0.447 |
Download Data |
Period Mean (original grids) [Pg] |
Model Period Mean (intersection) [Pg] |
Benchmark Period Mean (intersection) [Pg] |
Model Period Mean (complement) [Pg] |
Benchmark Period Mean (complement) [Pg] |
Bias [kg m-2] |
Bias Score [1] |
Spatial Distribution Score [1] |
Overall Score [1] |
|||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Benchmark | [-] | 8.47 | ||||||||||
| I123 | [-] | 5.81 | 5.76 | 8.47 | 0.129 | 7.29e-05 | -0.483 | 0.327 | 0.774 | 0.551 | ||
| post5.4 | [-] | 6.04 | 5.99 | 8.47 | 0.134 | 7.29e-05 | -0.418 | 0.324 | 0.795 | 0.559 |
Temporally integrated period mean