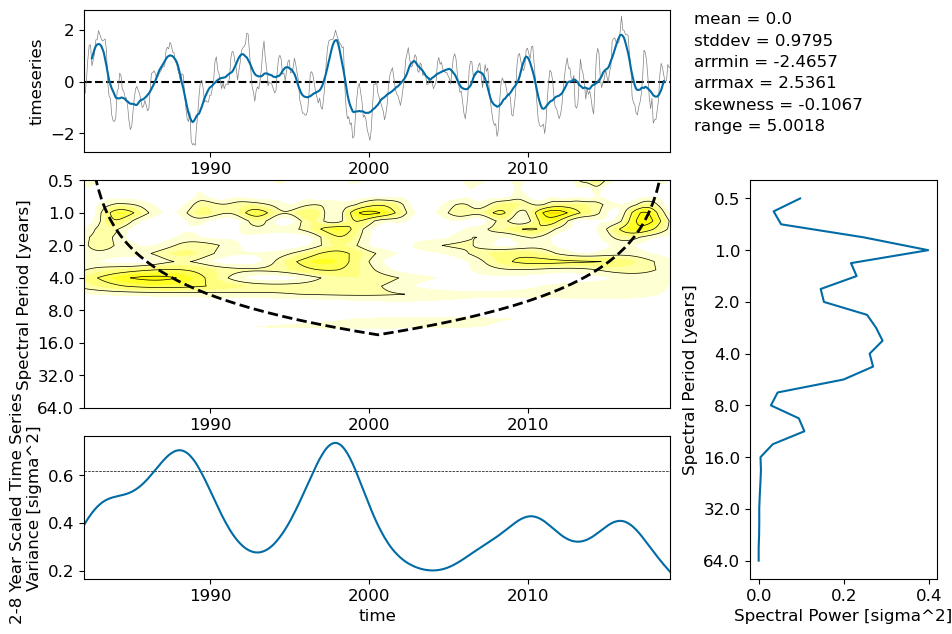
ENSO#
Show code cell source
%load_ext autoreload
%autoreload 2
Show code cell source
%%capture
# comment above line to see details about the run(s) displayed
from misc import *
from mom6_tools.enso import plot_enso_obs
import pickle
import cftime
import nc_time_axis
%matplotlib inline
Show code cell source
for path, case, i in zip(ocn_path, casename, range(len(casename))):
ds = xr.open_dataset(path+case+'_nino34_index.nc')
plot_enso(ds,label[i])
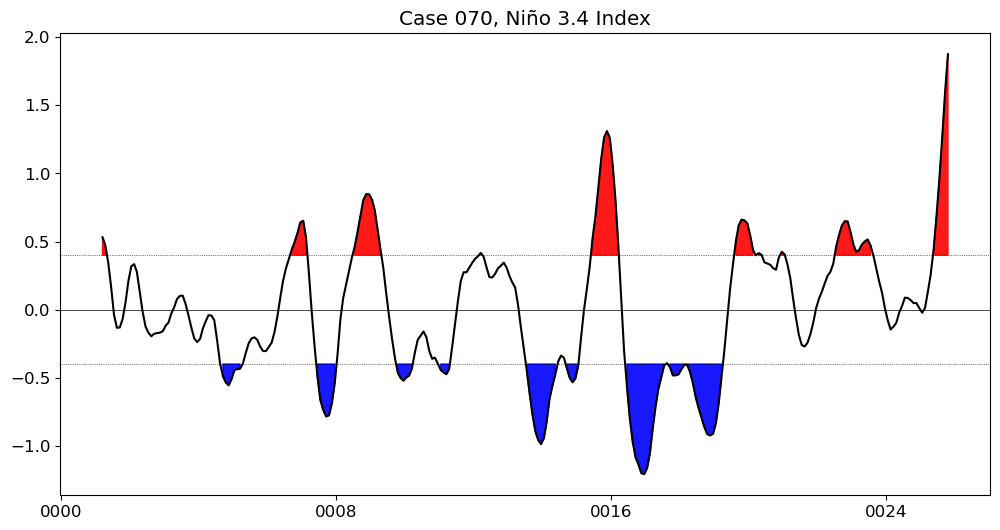
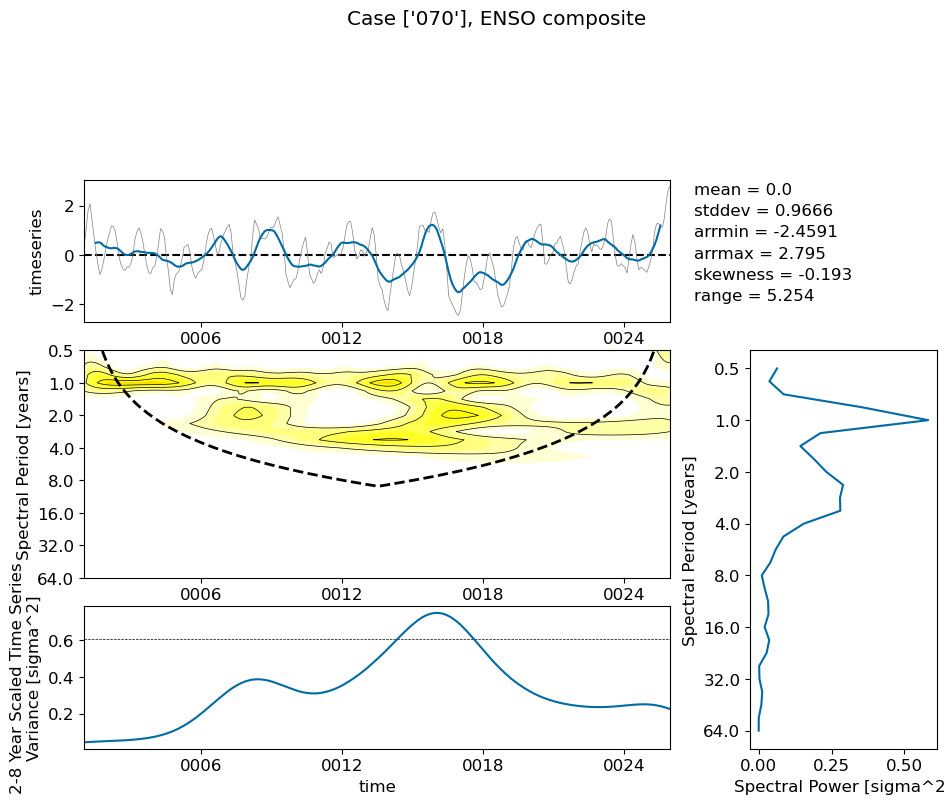
Show code cell source
for path, case, i in zip(ocn_path, casename, range(len(casename))):
fname = path+case+'_nino34_composite.pkl'
with open(fname, "rb") as file:
loaded_obj = pickle.load(file)
fig = loaded_obj.composite()
plt.suptitle('Case {}, ENSO composite'.format(label));
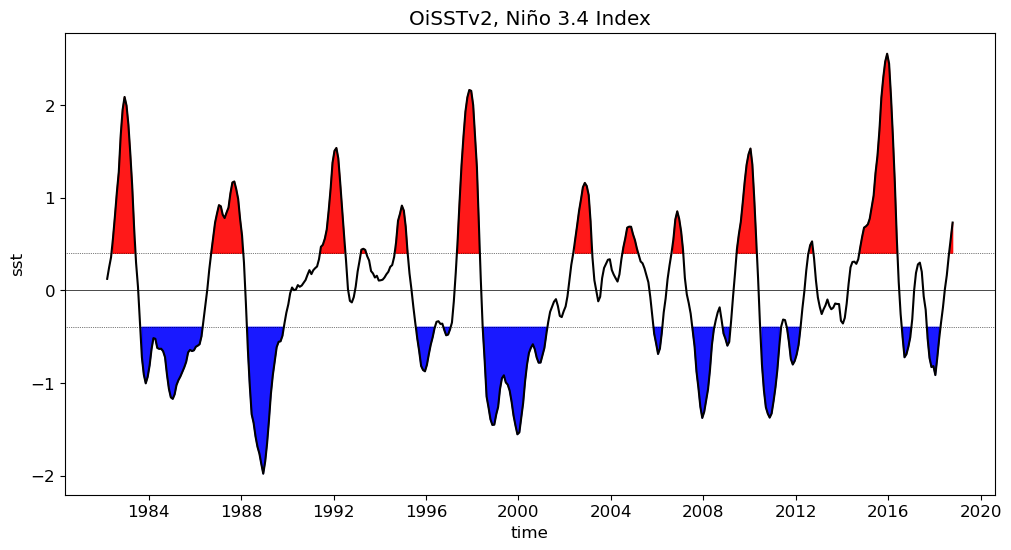
OiSSTv2#
Show code cell source
obs = xr.open_dataset('/glade/work/gmarques/cesm/tx2_3/oisst/oisstv2_to_tx2_3v2.nc')
#obs
Show code cell source
# Add the areacellonew coordinate
obs = obs.assign_coords({
"areacello": (("yh", "xh"), grd_xr[0].areacello.fillna(0.).data)
})
#obs
Show code cell source
plot_enso_obs(obs)