# Parameters
variable = "oml_min"
long_name = "Surface boundary layer depth"
oml_min#
from IPython.display import display, Markdown
# Dynamically generate markdown content
markdown_text = f" This notebook compares area-weighted mean and, in some cases, integral time series for {variable} in different basins."
# Display the updated markdown content
display(Markdown(markdown_text))
This notebook compares area-weighted mean and, in some cases, integral time series for oml_min in different basins.
%load_ext autoreload
%autoreload 2
%%capture
# comment above line to see details about the run(s) displayed
import sys, os
sys.path.append(os.path.abspath(".."))
from misc import *
import glob
print("Last update:", date.today())
%matplotlib inline
# figure size
fs = (10,4)
# load data
ds = []
for c, p in zip(casename, ocn_path):
file = glob.glob(p+'{}.native.{}.??????-??????.nc'.format(c, variable))[0]
ds.append(xr.open_dataset(file))
def ts_plot(variable, ds, fs, label, reg='Global'):
"""
Plot time series of regional means and integrals for a given variable from a list of datasets.
Parameters
----------
variable : str
Name of the variable to plot (prefix for "_mean" and "_int" variables in dataset).
ds : list of xarray.Dataset
List of datasets, each containing time series data for the specified variable with
variables named as `<variable>_mean` and optionally `<variable>_int`, and with
attributes 'long_name', 'units_mean', and optionally 'units_int'.
fs : tuple
Figure size (width, height) in inches for the plots.
label : list of str
List of labels corresponding to each dataset, used for the legend.
reg : str, optional
Name of the region to select for plotting (default is 'Global').
Returns
-------
None
Displays the plots but does not return any value.
Notes
-----
- This function creates one or two plots:
1. A time series of the variable's regional mean (`<variable>_mean`).
2. If available, a time series of the variable's regional integral (`<variable>_int`).
- The function expects each dataset to have attributes 'long_name', 'units_mean', and optionally 'units_int'.
- The same region name is applied across all datasets.
"""
fig, ax = plt.subplots(nrows=1, ncols=1, figsize=fs)
for l, i in zip(label, range(len(label))):
ds[i][variable+"_mean"].sel(region=reg).plot(ax=ax, label=l, lw=3, linestyle=linestyle[i], color=color[i])
long_name = ds[0].attrs['long_name']
ax.set_title("{}, {}".format(reg, long_name))
ax.set_ylabel(variable+"_mean, " + ds[i].attrs['units_mean'])
ax.set_xlabel('Year')
ax.grid()
ax.legend(ncol=3, loc=1)
if variable+"_int" in ds[0]:
fig, ax = plt.subplots(nrows=1, ncols=1, figsize=fs)
for l, i in zip(label, range(len(label))):
ds[i][variable+"_int"].sel(region=reg).plot(ax=ax, label=l, lw=3, linestyle=linestyle[i], color=color[i])
ax.set_title("{}, {}".format(reg, long_name))
ax.set_ylabel(variable+"_int, " + ds[i].attrs['units_int'])
ax.set_xlabel('Year')
ax.grid()
ax.legend(ncol=3, loc=1)
return
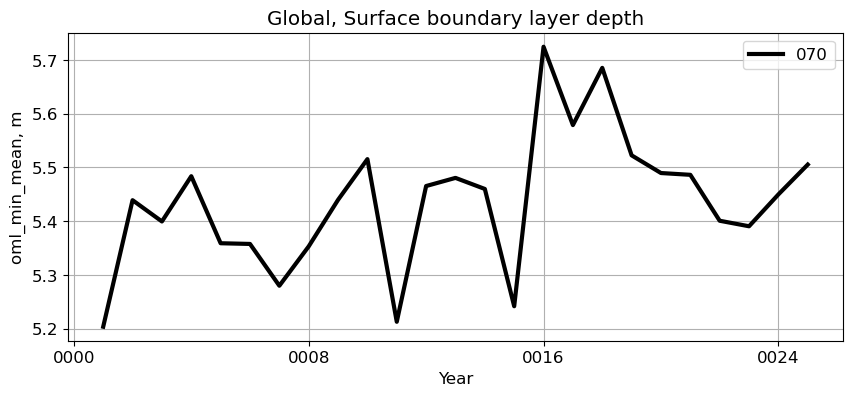
Global#
reg = 'Global'
ts_plot(variable, ds, fs, label, reg = reg)
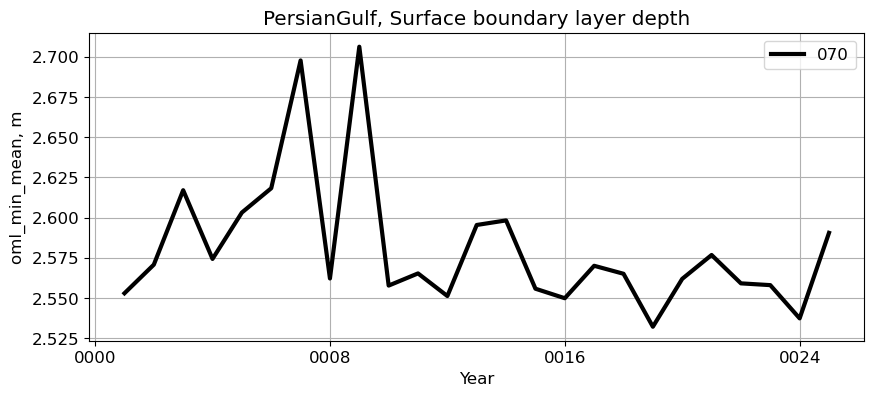
PersianGulf#
reg = 'PersianGulf'
ts_plot(variable, ds, fs, label, reg = reg)
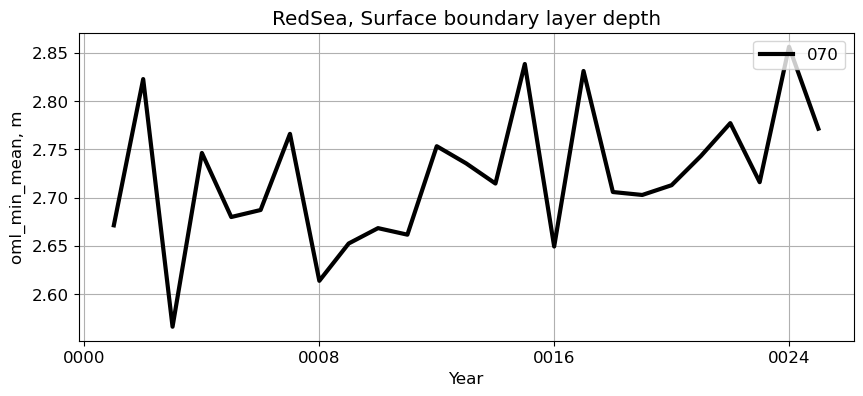
RedSea#
reg = 'RedSea'
ts_plot(variable, ds, fs, label, reg = reg)
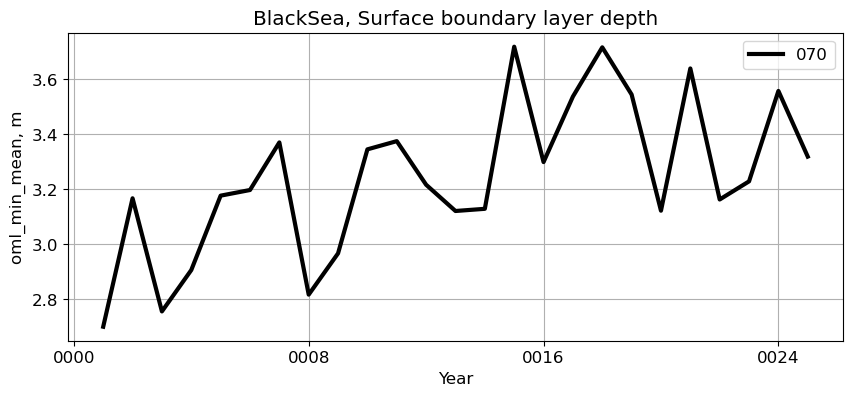
BlackSea#
reg = 'BlackSea'
ts_plot(variable, ds, fs, label, reg = reg)
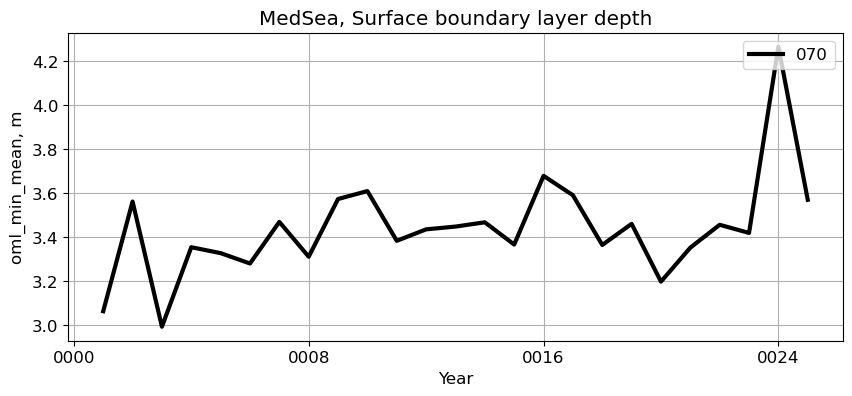
MedSea#
reg = 'MedSea'
ts_plot(variable, ds, fs, label, reg = reg)
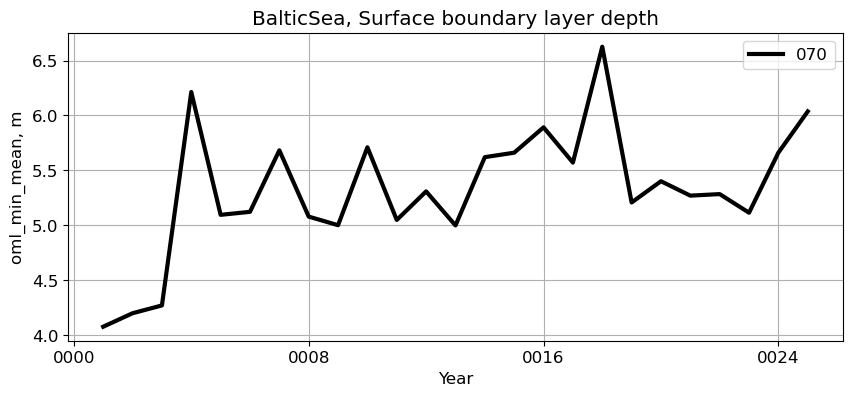
BalticSea#
reg = 'BalticSea'
ts_plot(variable, ds, fs, label, reg = reg)
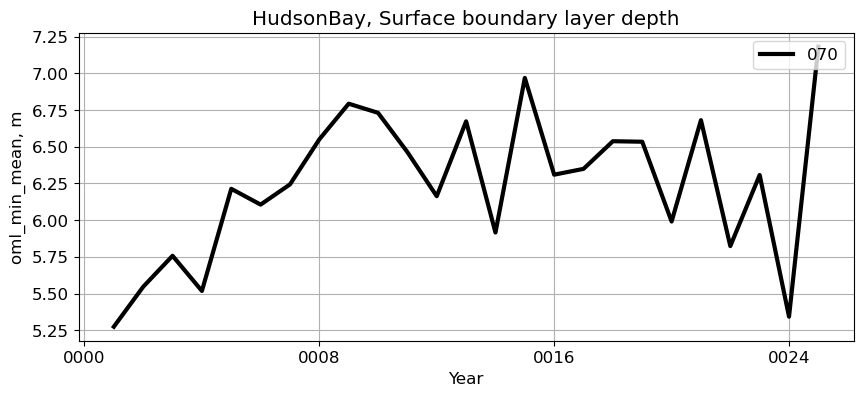
HudsonBay#
reg = 'HudsonBay'
ts_plot(variable, ds, fs, label, reg = reg)
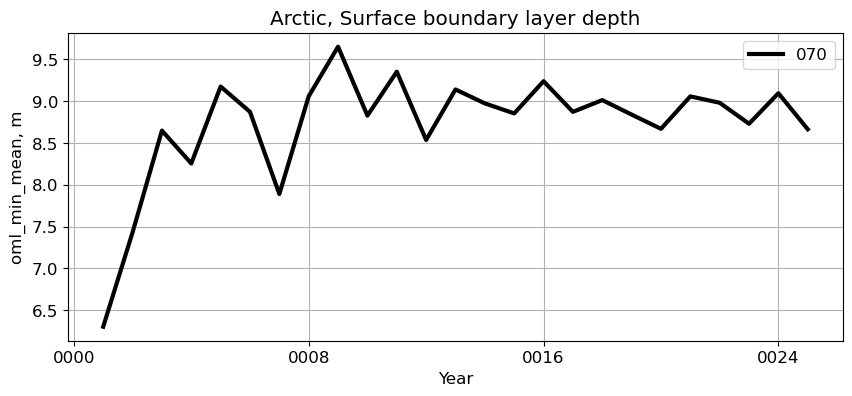
Arctic#
reg = 'Arctic'
ts_plot(variable, ds, fs, label, reg = reg)
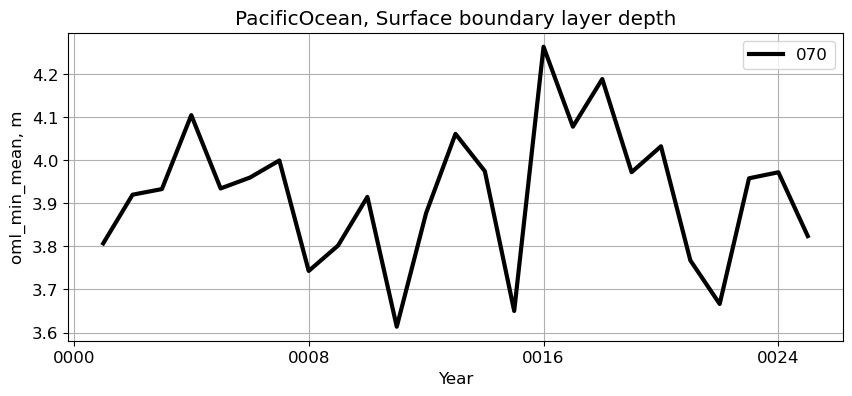
PacificOcean#
reg = 'PacificOcean'
ts_plot(variable, ds, fs, label, reg = reg)
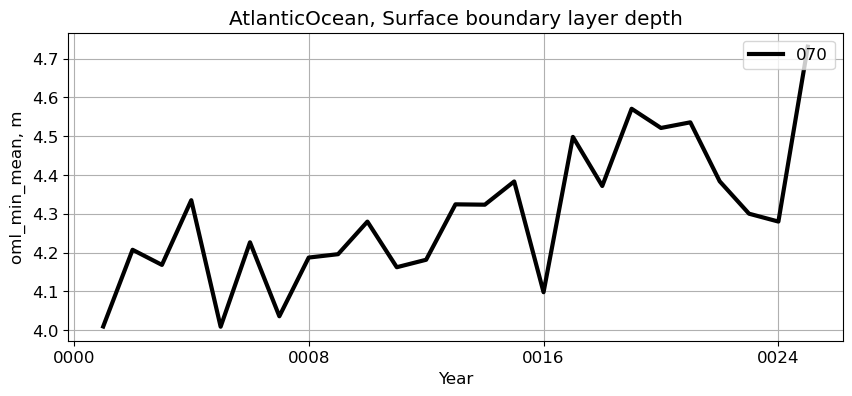
AtlanticOcean#
reg = 'AtlanticOcean'
ts_plot(variable, ds, fs, label, reg = reg)
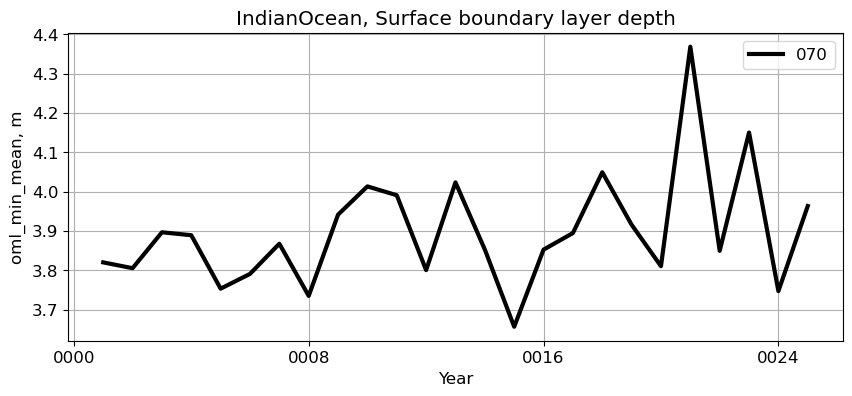
IndianOcean#
reg = 'IndianOcean'
ts_plot(variable, ds, fs, label, reg = reg)
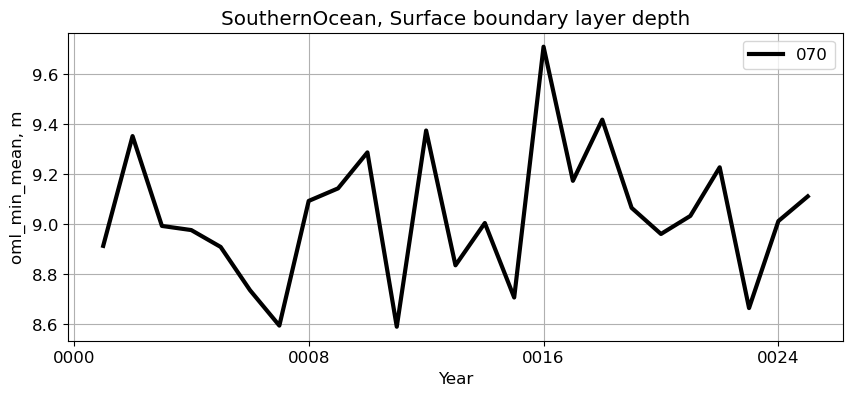
SouthernOcean#
reg = 'SouthernOcean'
ts_plot(variable, ds, fs, label, reg = reg)
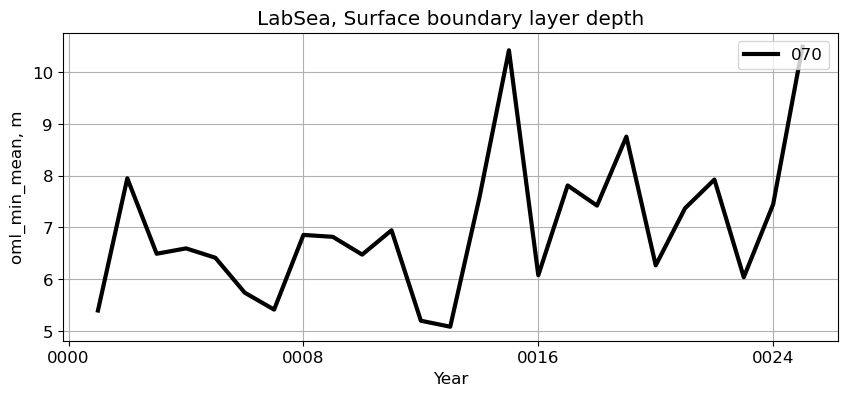
LabSea#
reg = 'LabSea'
ts_plot(variable, ds, fs, label, reg = reg)
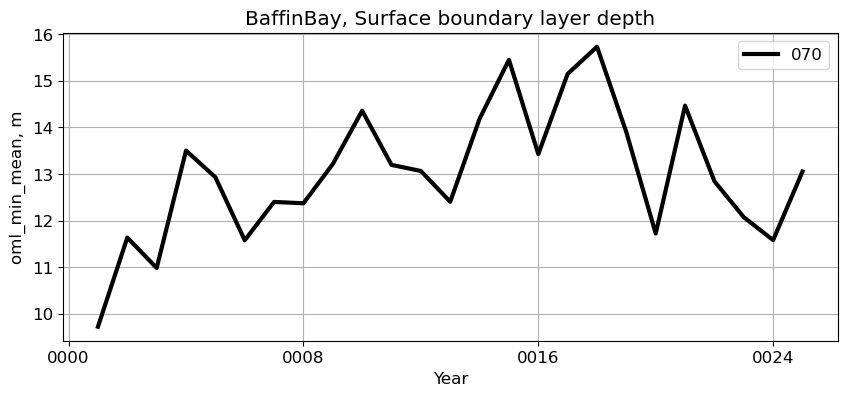
BaffinBay#
reg = 'BaffinBay'
ts_plot(variable, ds, fs, label, reg = reg)
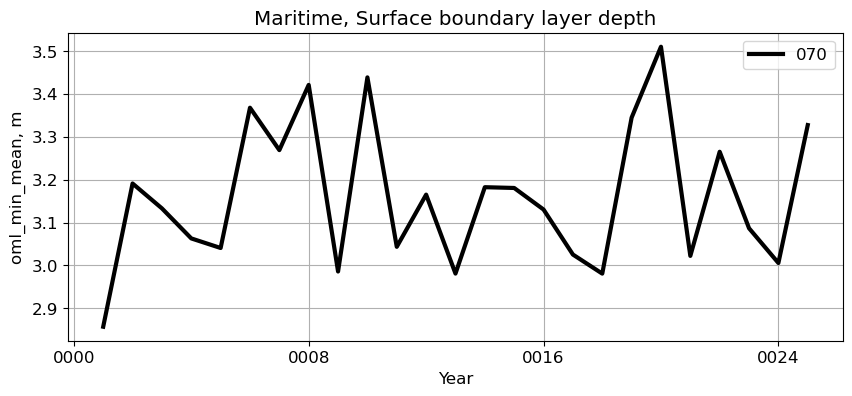
Maritime#
reg = 'Maritime'
ts_plot(variable, ds, fs, label, reg = reg)
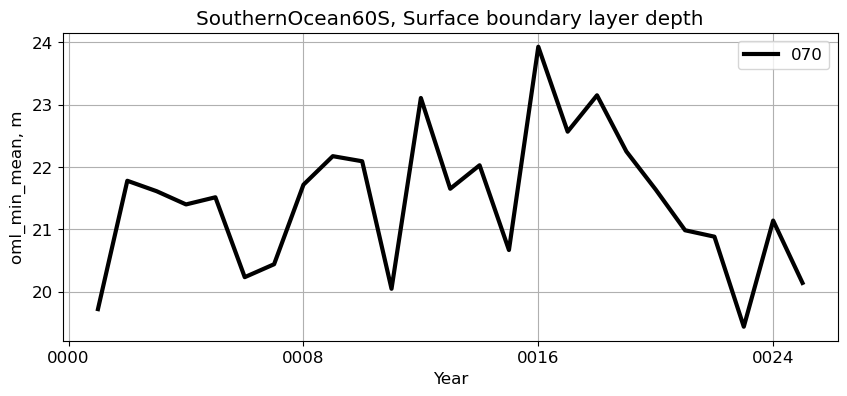
SouthernOcean60S#
reg = 'SouthernOcean60S'
ts_plot(variable, ds, fs, label, reg = reg)
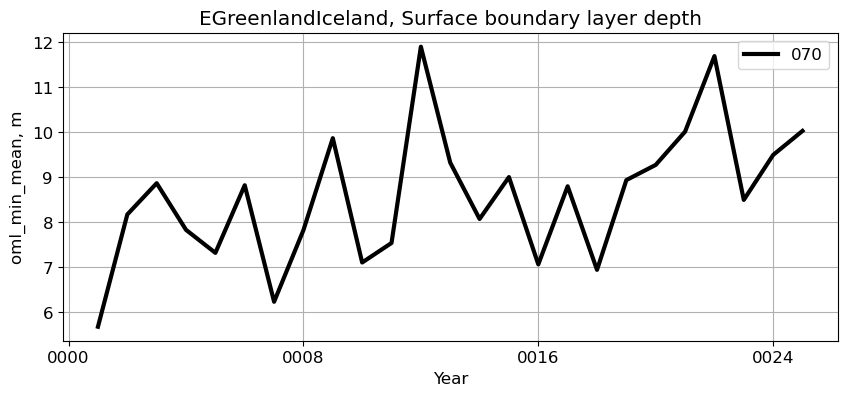
EGreenlandIceland#
reg = 'EGreenlandIceland'
ts_plot(variable, ds, fs, label, reg = reg)
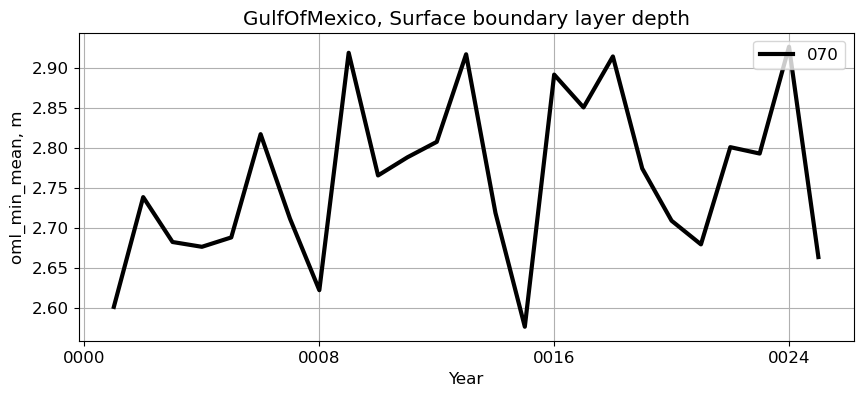
GulfOfMexico#
reg = 'GulfOfMexico'
ts_plot(variable, ds, fs, label, reg = reg)