Mixed layer depth#
%load_ext autoreload
%autoreload 2
%%capture
# comment above line to see details about the run(s) displayed
from misc import *
from mom6_tools.m6plot import myStats, annotateStats, xycompare
import cartopy.crs as ccrs
import cartopy.feature
import intake
%matplotlib inline
# load obs-based mld from oce-catalog
obs = 'mld-deboyer-tx2_3v2'
catalog = intake.open_catalog(diag_config_yml['oce_cat'])
print('\n Reading climatology from: ', obs)
mld_obs = catalog[obs].to_dask().where(grd_xr[0].wet > 0.)
# rename coords and and fix time
mld_obs = mld_obs.rename({'lon' : 'longitude', 'lat' : 'latitude', 'time' : 'month'})
mld_obs["month"] = np.arange(1,len(mld_obs.month)+1)
months = [0,1,2]
obs_JFM = np.ma.masked_invalid(mld_obs.mld.isel(month=months).mean('month').values)
months = [6,7,8]
obs_JAS = np.ma.masked_invalid(mld_obs.mld.isel(month=months).mean('month').values)
obs_winter = obs_JAS.copy(); obs_summer = obs_JAS.copy()
j = np.abs( grd[0].geolat[:,0] - 0. ).argmin()
obs_winter[j::,:] = obs_JFM[j::,:]
obs_summer[0:j,:] = obs_JFM[0:j,:]
Reading climatology from: mld-deboyer-tx2_3v2
Monthly#
months = ['January', 'February', 'March', 'April',
'May', 'June', 'July', 'August', 'September',
'October', 'November', 'December']
ds = []
for path, case, i in zip(ocn_path, casename, range(len(label))):
ds.append(xr.open_dataset(path+case+'_MLD_monthly_clima.nc'))
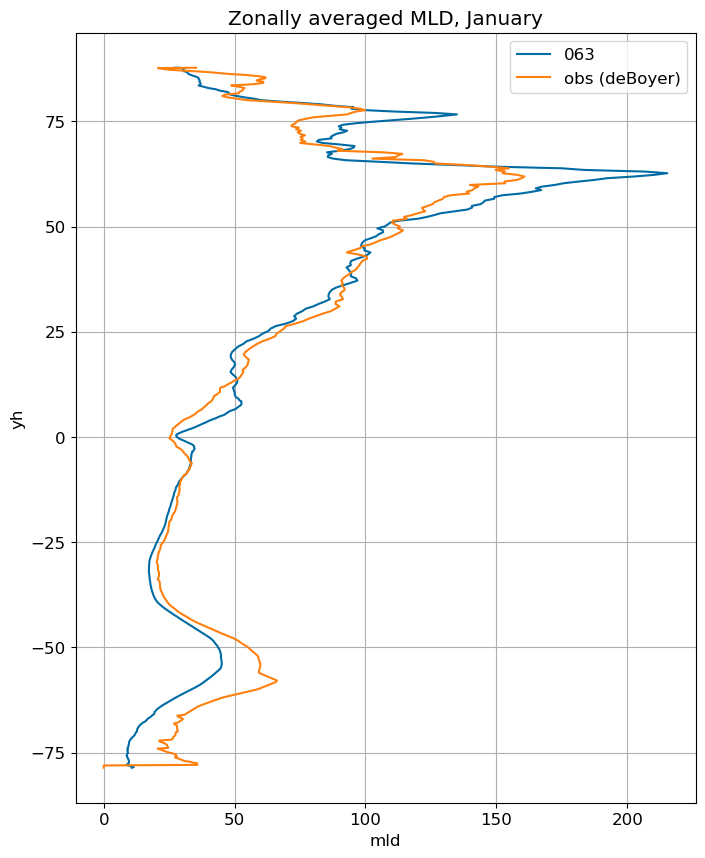
January#
m=0
for i in range(len(label)):
model = np.ma.masked_invalid(ds[i].mlotst.isel(month=m).values)
obs = np.ma.masked_where(grd[i].wet == 0, mld_obs.mld.isel(month=m).values)
xycompare(model,
obs,
grd[i].geolon, grd[i].geolat, grd[0].areacello,
title1 = label[i],
title2 = 'obs (deBoyer)',
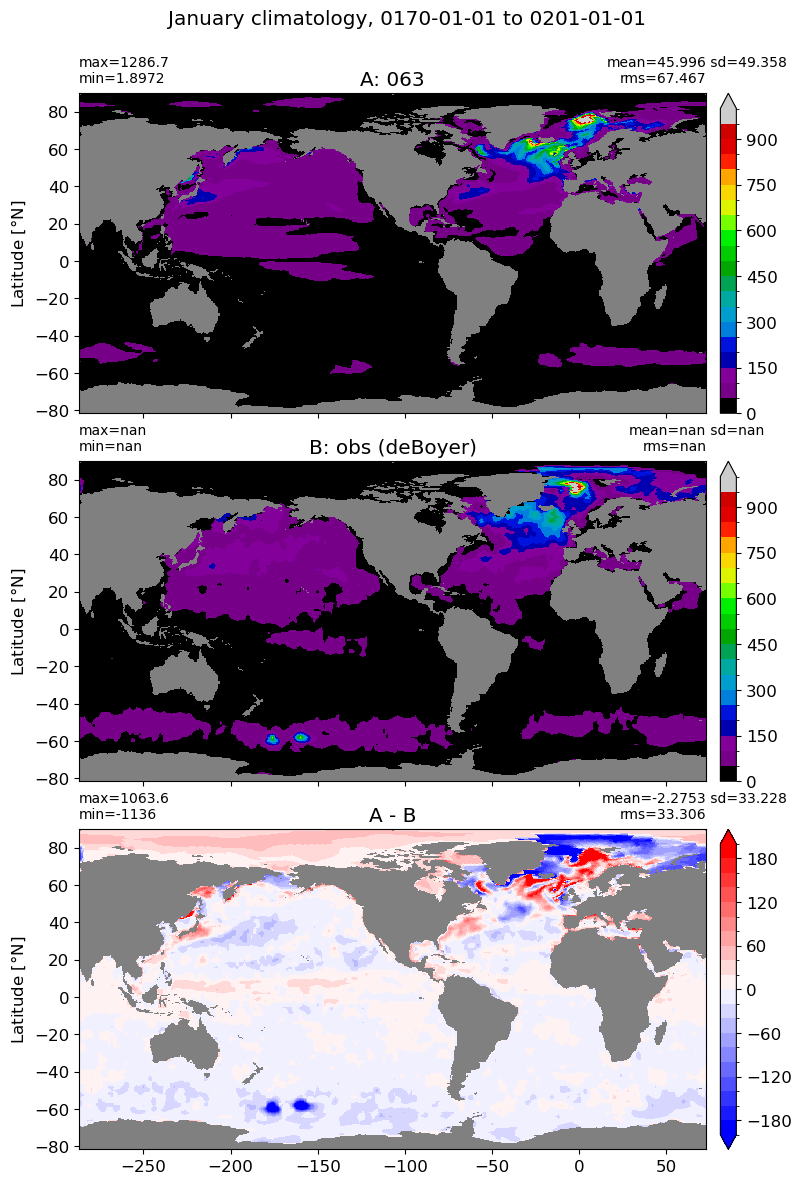
suptitle=str(months[m]) +' climatology, '+ str(start_date) + ' to ' + str(end_date),
colormap=plt.cm.nipy_spectral, dcolormap=plt.cm.bwr,
clim = (0,1000), extend='max',
dlim=(-200,200))
fig, ax = plt.subplots(figsize=(8,10))
for i in range(len(label)):
ds[i].mlotst.isel(month=m).weighted(grd_xr[i].areacello.fillna(0)).mean('xh').plot(y="yh",
ax=ax, label=label[i])
mld_obs.mld.isel(month=m).weighted(grd_xr[0].areacello.fillna(0)).mean('xh').plot(y="yh",
ax=ax, label='obs (deBoyer)')
ax.set_title('Zonally averaged MLD, '+str(months[m]))
ax.grid()
ax.legend();
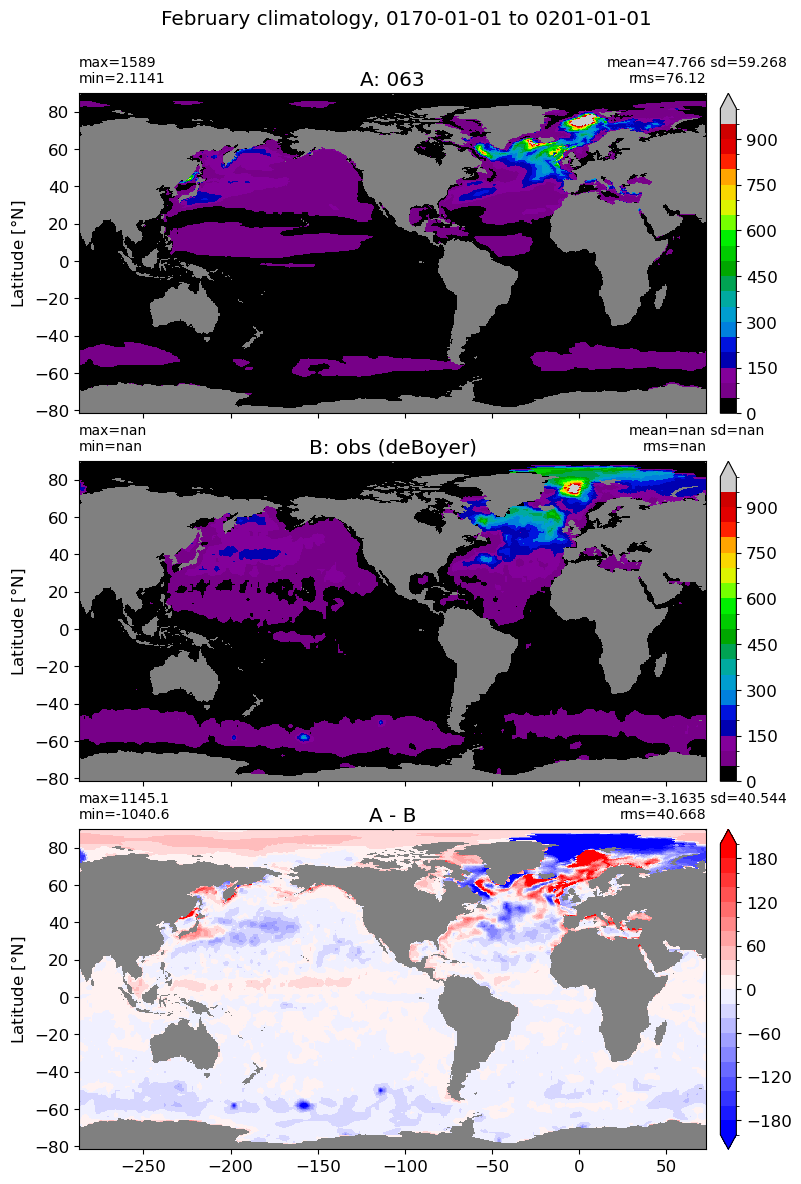
February#
m=1
for i in range(len(label)):
model = np.ma.masked_invalid(ds[i].mlotst.isel(month=m).values)
obs = np.ma.masked_where(grd[i].wet == 0, mld_obs.mld.isel(month=m).values)
xycompare(model,
obs,
grd[i].geolon, grd[i].geolat, grd[0].areacello,
title1 = label[i],
title2 = 'obs (deBoyer)',
suptitle=str(months[m]) +' climatology, '+ str(start_date) + ' to ' + str(end_date),
colormap=plt.cm.nipy_spectral, dcolormap=plt.cm.bwr,
clim = (0,1000), extend='max',
dlim=(-200,200))
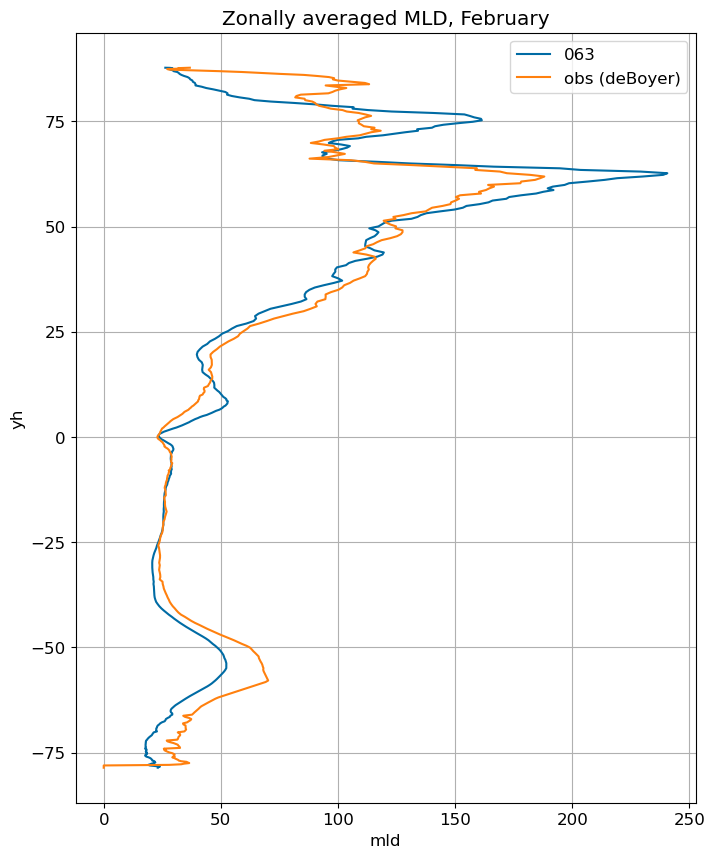
fig, ax = plt.subplots(figsize=(8,10))
for i in range(len(label)):
ds[i].mlotst.isel(month=m).weighted(grd_xr[i].areacello.fillna(0)).mean('xh').plot(y="yh",
ax=ax, label=label[i])
mld_obs.mld.isel(month=m).weighted(grd_xr[0].areacello.fillna(0)).mean('xh').plot(y="yh",
ax=ax, label='obs (deBoyer)')
ax.set_title('Zonally averaged MLD, '+str(months[m]))
ax.grid()
ax.legend();
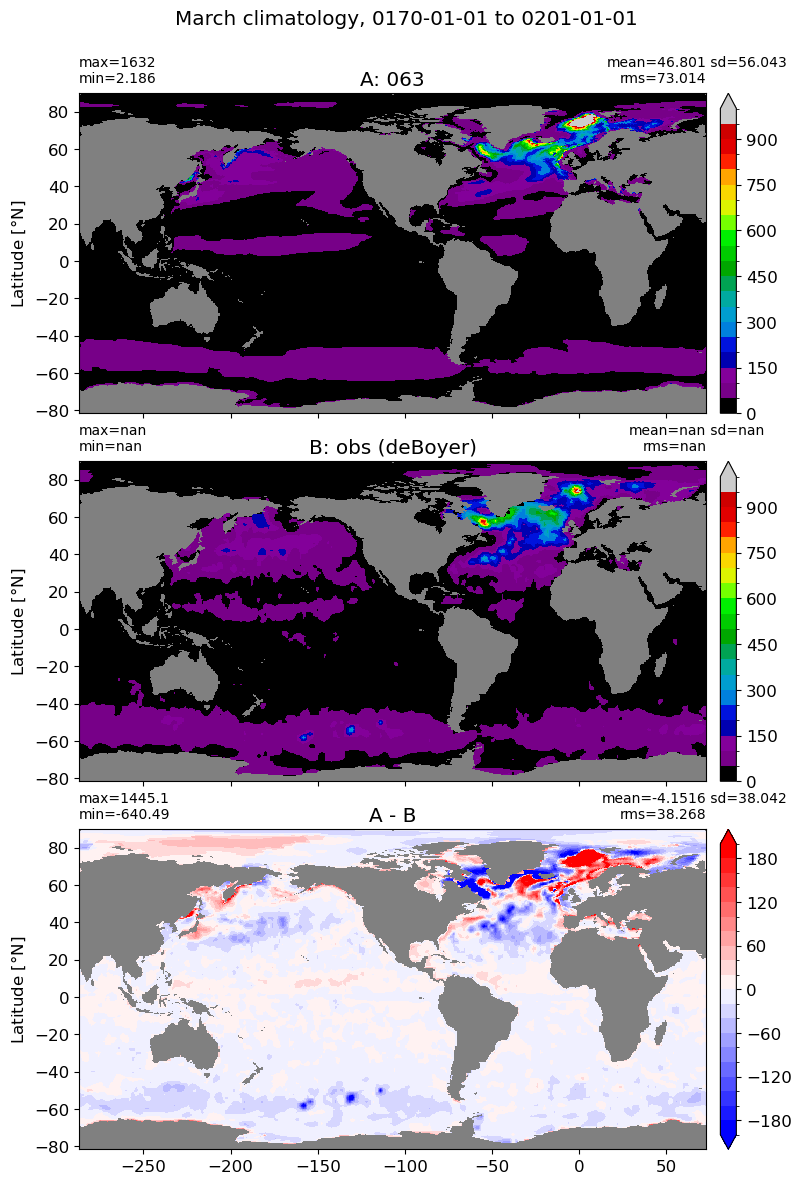
March#
m=2
for i in range(len(label)):
model = np.ma.masked_invalid(ds[i].mlotst.isel(month=m).values)
obs = np.ma.masked_where(grd[i].wet == 0, mld_obs.mld.isel(month=m).values)
xycompare(model,
obs,
grd[i].geolon, grd[i].geolat, grd[0].areacello,
title1 = label[i],
title2 = 'obs (deBoyer)',
suptitle=str(months[m]) +' climatology, '+ str(start_date) + ' to ' + str(end_date),
colormap=plt.cm.nipy_spectral, dcolormap=plt.cm.bwr,
clim = (0,1000), extend='max',
dlim=(-200,200))
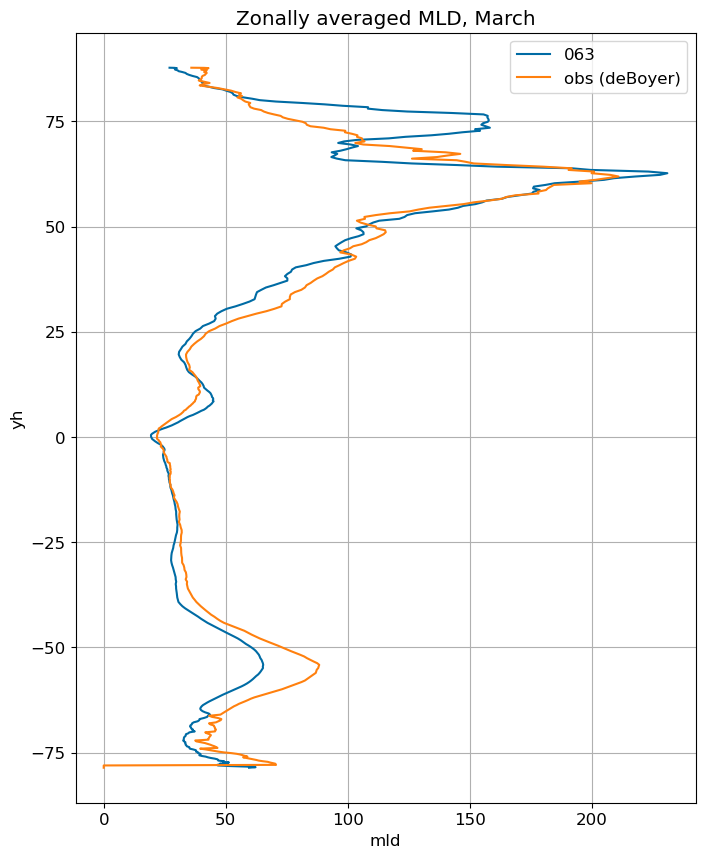
fig, ax = plt.subplots(figsize=(8,10))
for i in range(len(label)):
ds[i].mlotst.isel(month=m).weighted(grd_xr[i].areacello.fillna(0)).mean('xh').plot(y="yh",
ax=ax, label=label[i])
mld_obs.mld.isel(month=m).weighted(grd_xr[0].areacello.fillna(0)).mean('xh').plot(y="yh",
ax=ax, label='obs (deBoyer)')
ax.set_title('Zonally averaged MLD, '+str(months[m]))
ax.grid()
ax.legend();
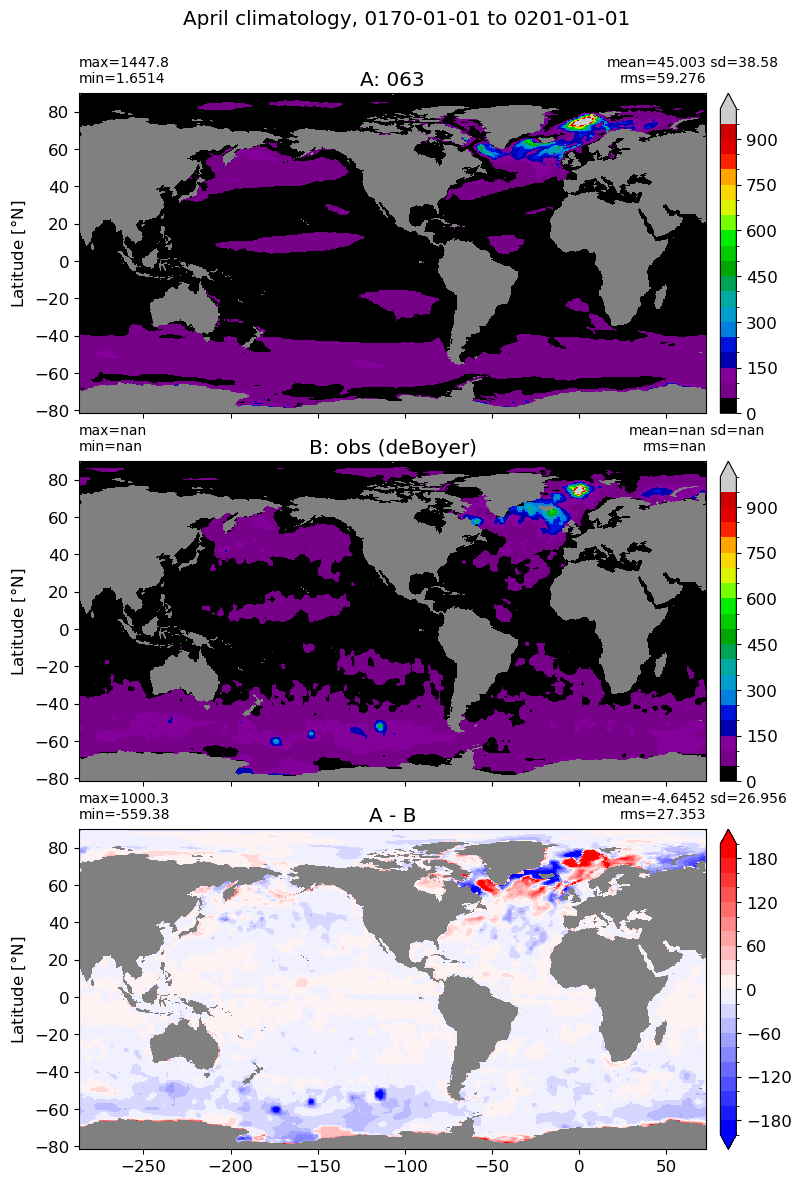
April#
m=3
for i in range(len(label)):
model = np.ma.masked_invalid(ds[i].mlotst.isel(month=m).values)
obs = np.ma.masked_where(grd[i].wet == 0, mld_obs.mld.isel(month=m).values)
xycompare(model,
obs,
grd[i].geolon, grd[i].geolat, grd[0].areacello,
title1 = label[i],
title2 = 'obs (deBoyer)',
suptitle=str(months[m]) +' climatology, '+ str(start_date) + ' to ' + str(end_date),
colormap=plt.cm.nipy_spectral, dcolormap=plt.cm.bwr,
clim = (0,1000), extend='max',
dlim=(-200,200))
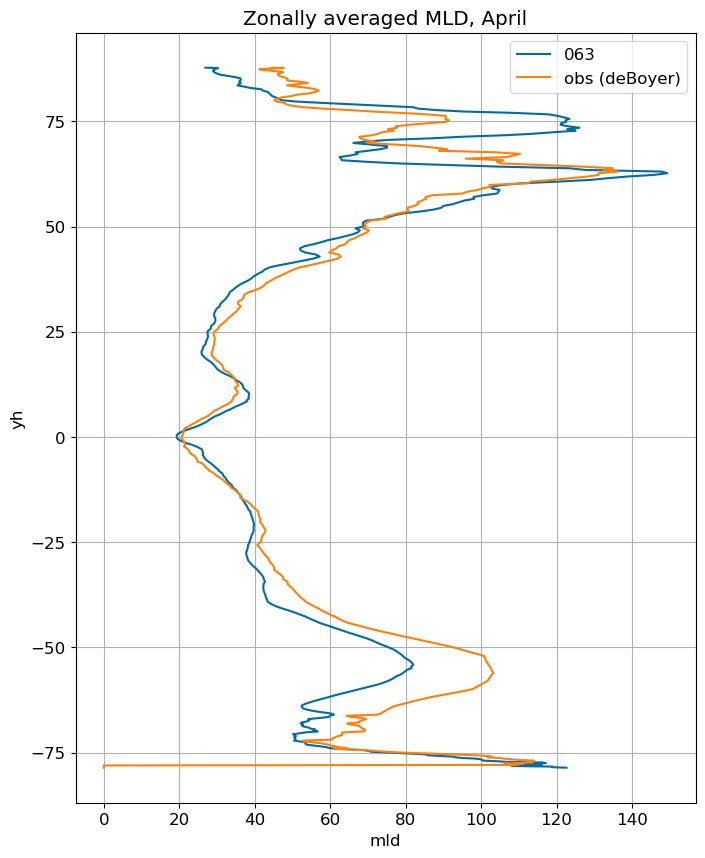
fig, ax = plt.subplots(figsize=(8,10))
for i in range(len(label)):
ds[i].mlotst.isel(month=m).weighted(grd_xr[i].areacello.fillna(0)).mean('xh').plot(y="yh",
ax=ax, label=label[i])
mld_obs.mld.isel(month=m).weighted(grd_xr[0].areacello.fillna(0)).mean('xh').plot(y="yh",
ax=ax, label='obs (deBoyer)')
ax.set_title('Zonally averaged MLD, '+str(months[m]))
ax.grid()
ax.legend();
May#
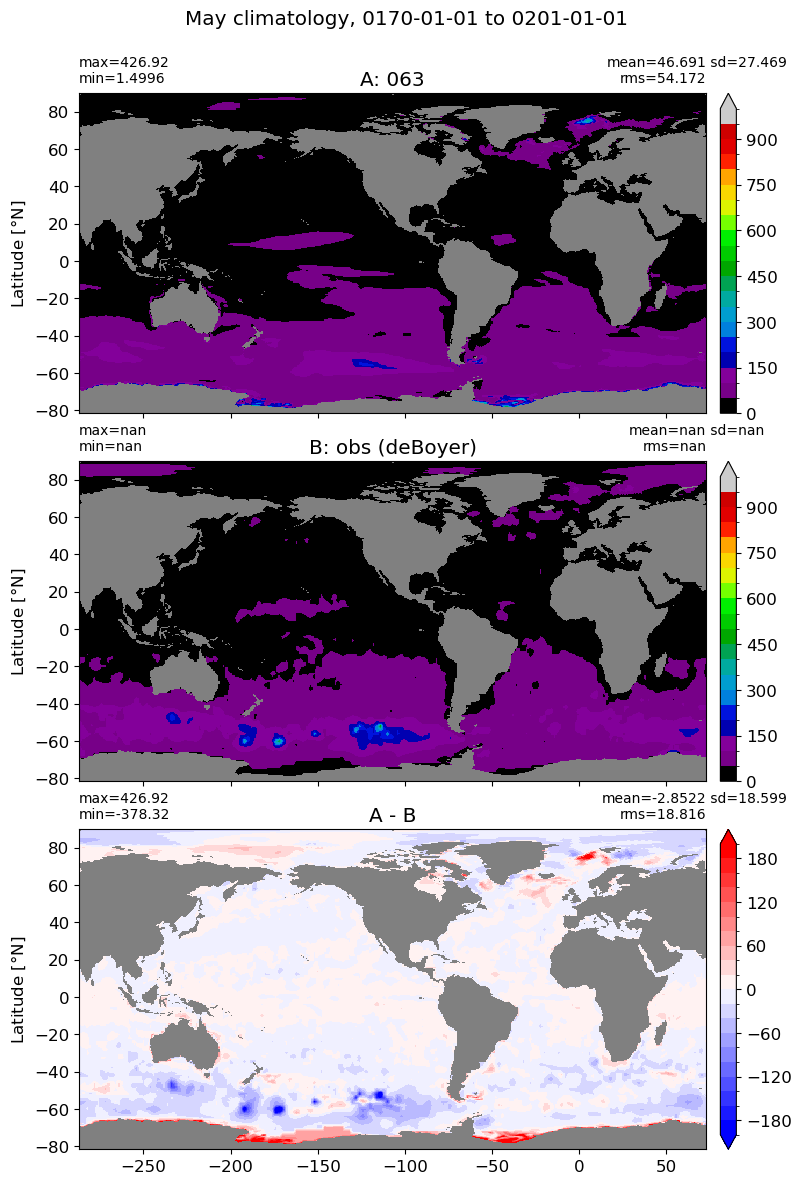
m=4
for i in range(len(label)):
model = np.ma.masked_invalid(ds[i].mlotst.isel(month=m).values)
obs = np.ma.masked_where(grd[i].wet == 0, mld_obs.mld.isel(month=m).values)
xycompare(model,
obs,
grd[i].geolon, grd[i].geolat, grd[0].areacello,
title1 = label[i],
title2 = 'obs (deBoyer)',
suptitle=str(months[m]) +' climatology, '+ str(start_date) + ' to ' + str(end_date),
colormap=plt.cm.nipy_spectral, dcolormap=plt.cm.bwr,
clim = (0,1000), extend='max',
dlim=(-200,200))
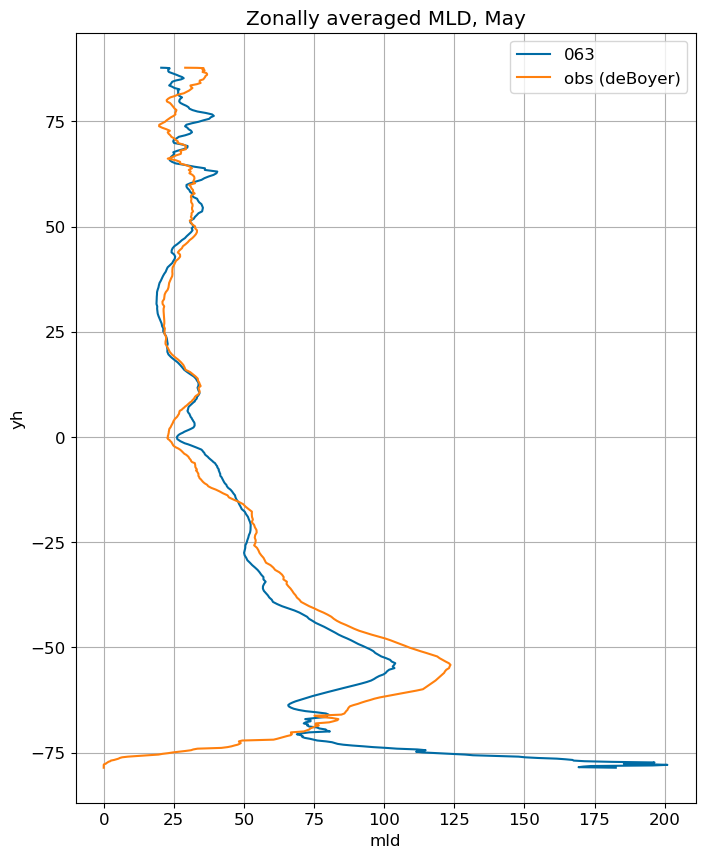
fig, ax = plt.subplots(figsize=(8,10))
for i in range(len(label)):
ds[i].mlotst.isel(month=m).weighted(grd_xr[i].areacello.fillna(0)).mean('xh').plot(y="yh",
ax=ax, label=label[i])
mld_obs.mld.isel(month=m).weighted(grd_xr[0].areacello.fillna(0)).mean('xh').plot(y="yh",
ax=ax, label='obs (deBoyer)')
ax.set_title('Zonally averaged MLD, '+str(months[m]))
ax.grid()
ax.legend();
June#
m=5
for i in range(len(label)):
model = np.ma.masked_invalid(ds[i].mlotst.isel(month=m).values)
obs = np.ma.masked_where(grd[i].wet == 0, mld_obs.mld.isel(month=m).values)
xycompare(model,
obs,
grd[i].geolon, grd[i].geolat, grd[0].areacello,
title1 = label[i],
title2 = 'obs (deBoyer)',
suptitle=str(months[m]) +' climatology, '+ str(start_date) + ' to ' + str(end_date),
colormap=plt.cm.nipy_spectral, dcolormap=plt.cm.bwr,
clim = (0,1000), extend='max',
dlim=(-200,200))
fig, ax = plt.subplots(figsize=(8,10))
for i in range(len(label)):
ds[i].mlotst.isel(month=m).mean('xh').plot(y="yh", ax=ax, label=label[i])
mld_obs.mld.isel(month=m).mean('xh').plot(y="yh", ax=ax, label='obs (deBoyer)')
ax.set_title('Zonally averaged MLD, '+str(months[m]))
ax.grid()
ax.legend();
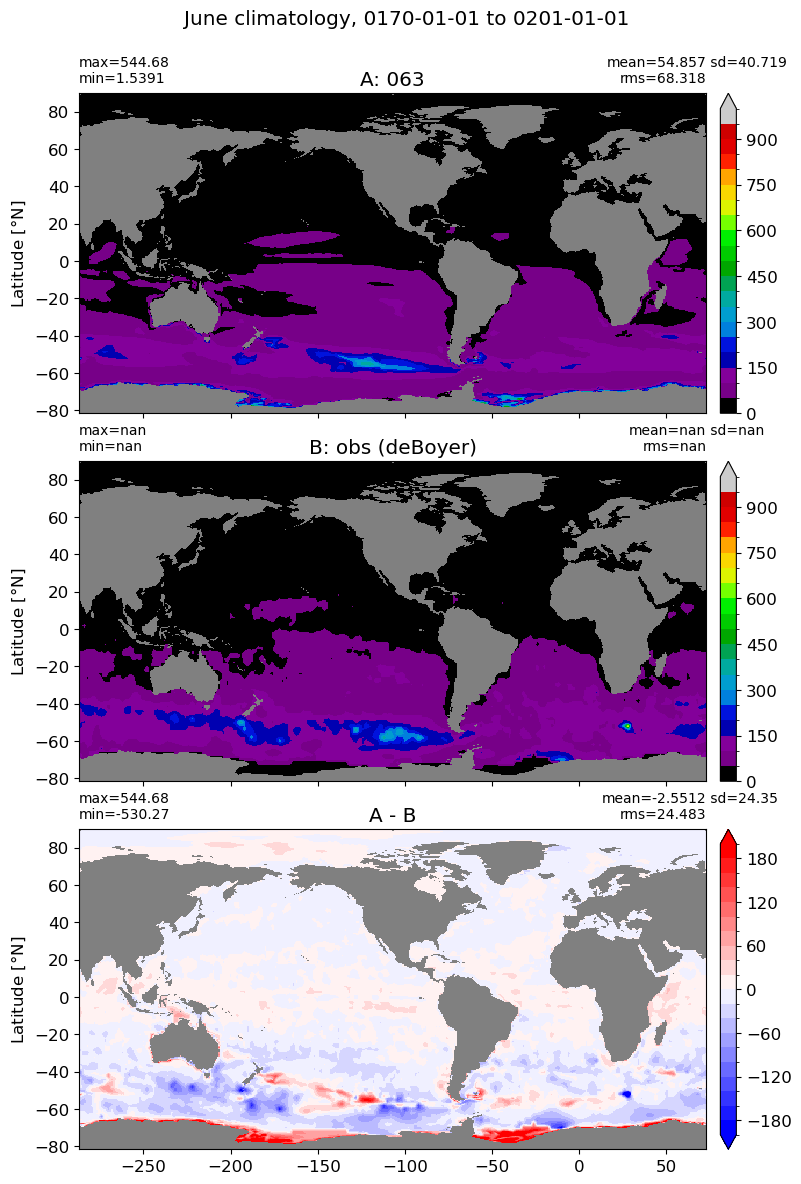
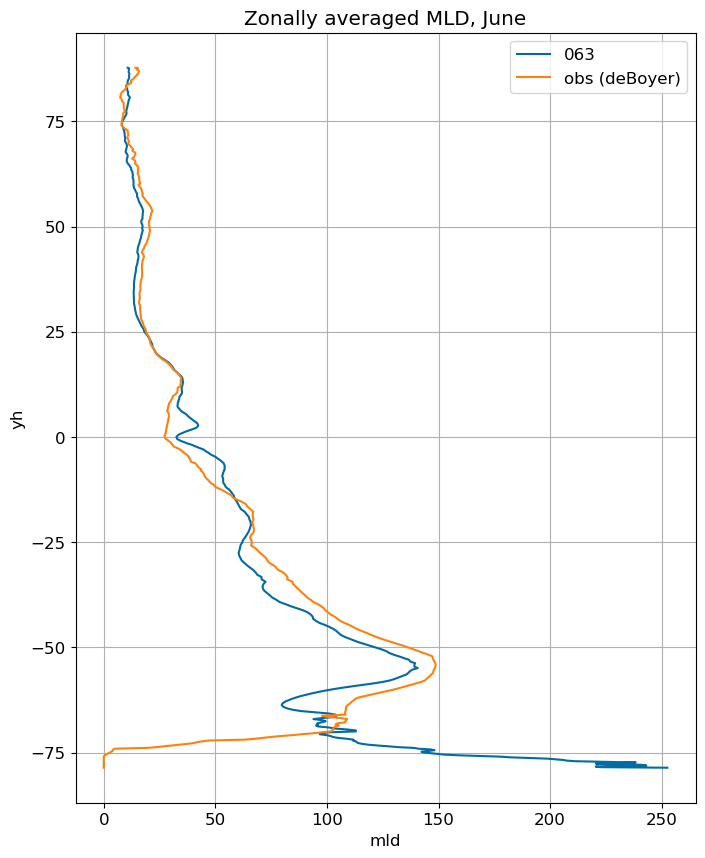
July#
m=6
for i in range(len(label)):
model = np.ma.masked_invalid(ds[i].mlotst.isel(month=m).values)
obs = np.ma.masked_where(grd[i].wet == 0, mld_obs.mld.isel(month=m).values)
xycompare(model,
obs,
grd[i].geolon, grd[i].geolat, grd[0].areacello,
title1 = label[i],
title2 = 'obs (deBoyer)',
suptitle=str(months[m]) +' climatology, '+ str(start_date) + ' to ' + str(end_date),
colormap=plt.cm.nipy_spectral, dcolormap=plt.cm.bwr,
clim = (0,1000), extend='max',
dlim=(-200,200))
fig, ax = plt.subplots(figsize=(8,10))
for i in range(len(label)):
ds[i].mlotst.isel(month=m).weighted(grd_xr[i].areacello.fillna(0)).mean('xh').plot(y="yh",
ax=ax, label=label[i])
mld_obs.mld.isel(month=m).weighted(grd_xr[0].areacello.fillna(0)).mean('xh').plot(y="yh",
ax=ax, label='obs (deBoyer)')
ax.set_title('Zonally averaged MLD, '+str(months[m]))
ax.grid()
ax.legend();
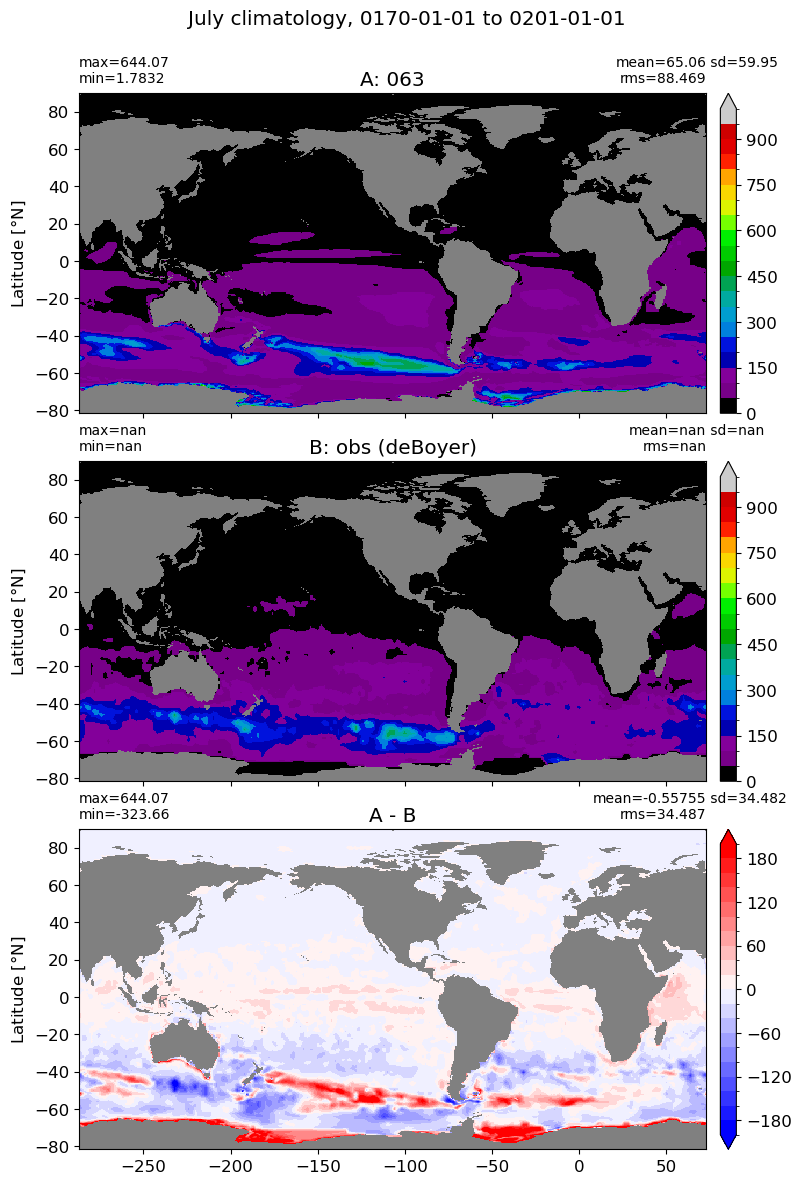
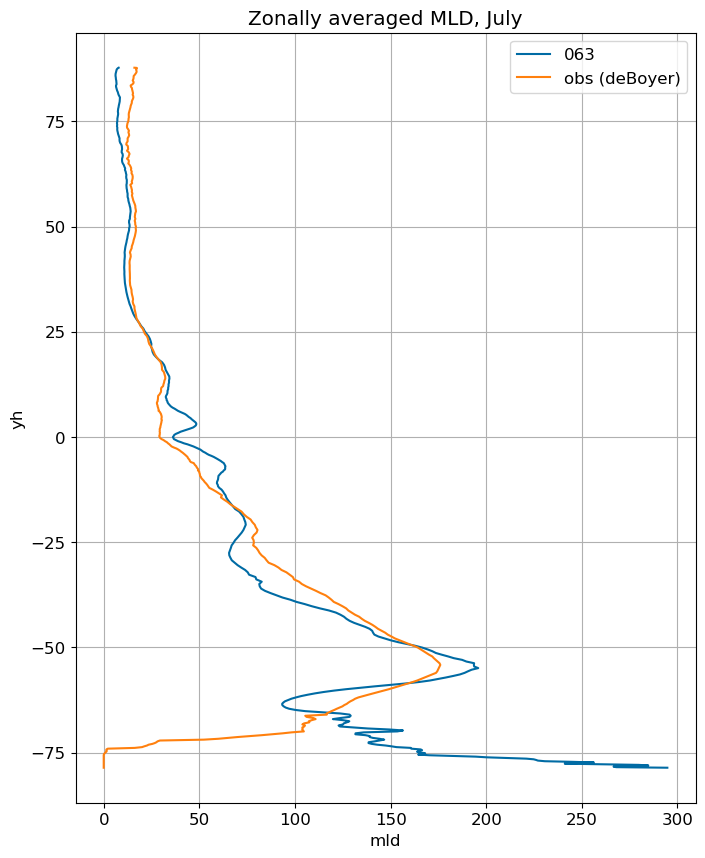
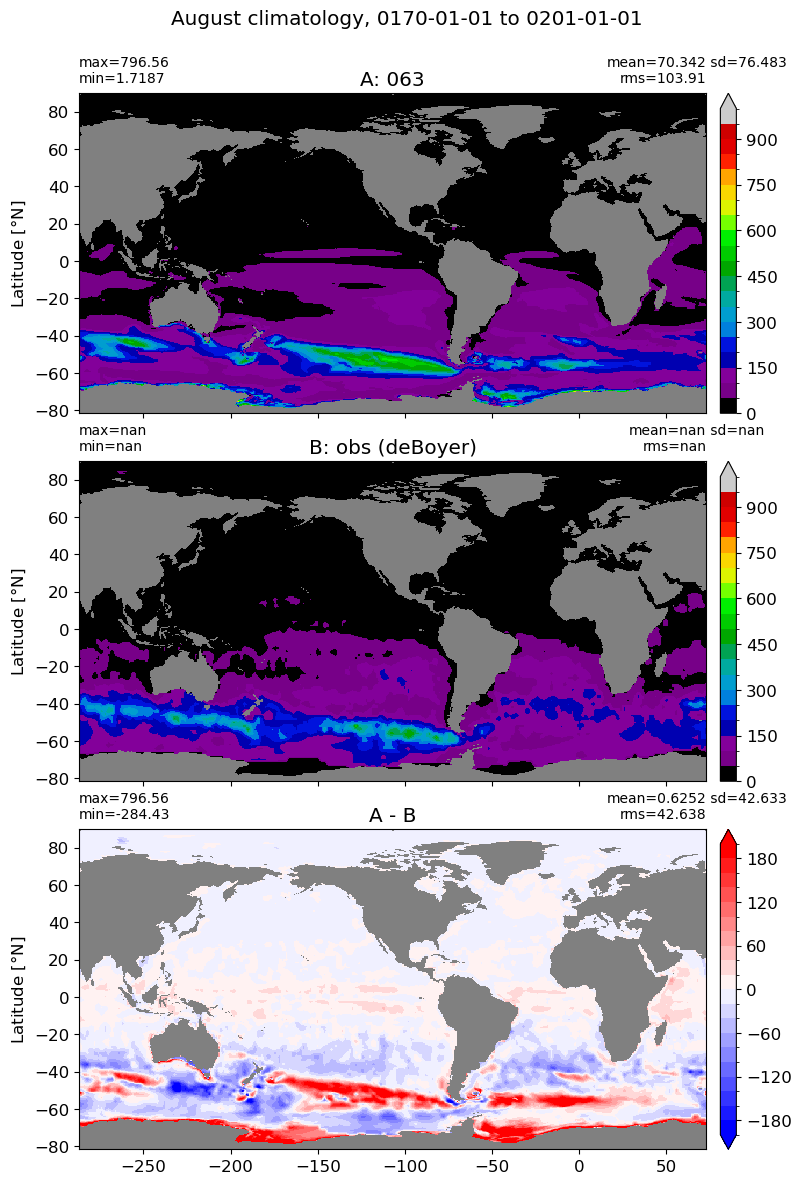
August#
m=7
for i in range(len(label)):
model = np.ma.masked_invalid(ds[i].mlotst.isel(month=m).values)
obs = np.ma.masked_where(grd[i].wet == 0, mld_obs.mld.isel(month=m).values)
xycompare(model,
obs,
grd[i].geolon, grd[i].geolat, grd[0].areacello,
title1 = label[i],
title2 = 'obs (deBoyer)',
suptitle=str(months[m]) +' climatology, '+ str(start_date) + ' to ' + str(end_date),
colormap=plt.cm.nipy_spectral, dcolormap=plt.cm.bwr,
clim = (0,1000), extend='max',
dlim=(-200,200))
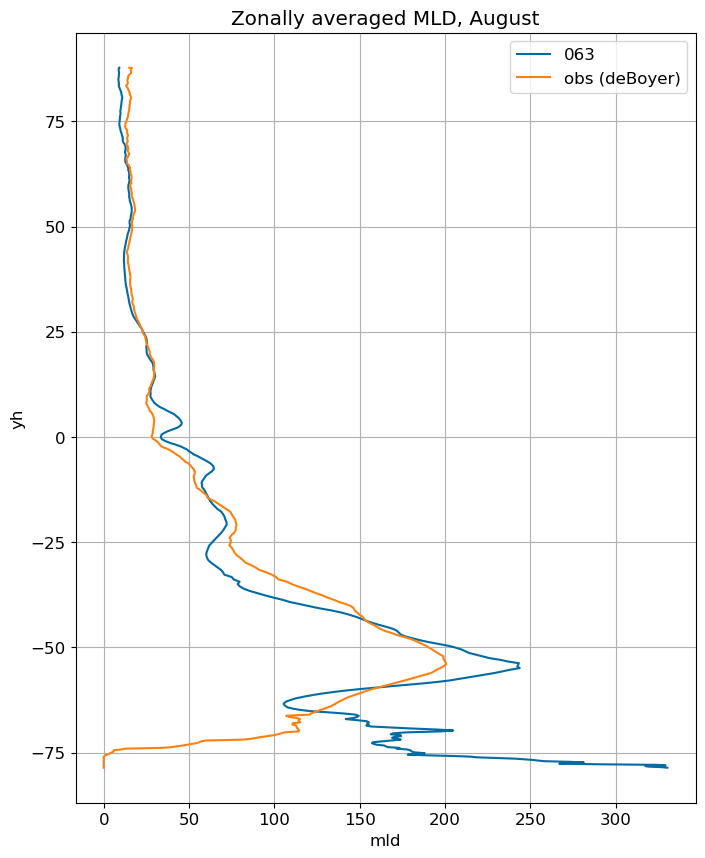
fig, ax = plt.subplots(figsize=(8,10))
for i in range(len(label)):
ds[i].mlotst.isel(month=m).weighted(grd_xr[i].areacello.fillna(0)).mean('xh').plot(y="yh",
ax=ax, label=label[i])
mld_obs.mld.isel(month=m).weighted(grd_xr[0].areacello.fillna(0)).mean('xh').plot(y="yh",
ax=ax, label='obs (deBoyer)')
ax.set_title('Zonally averaged MLD, '+str(months[m]))
ax.grid()
ax.legend();
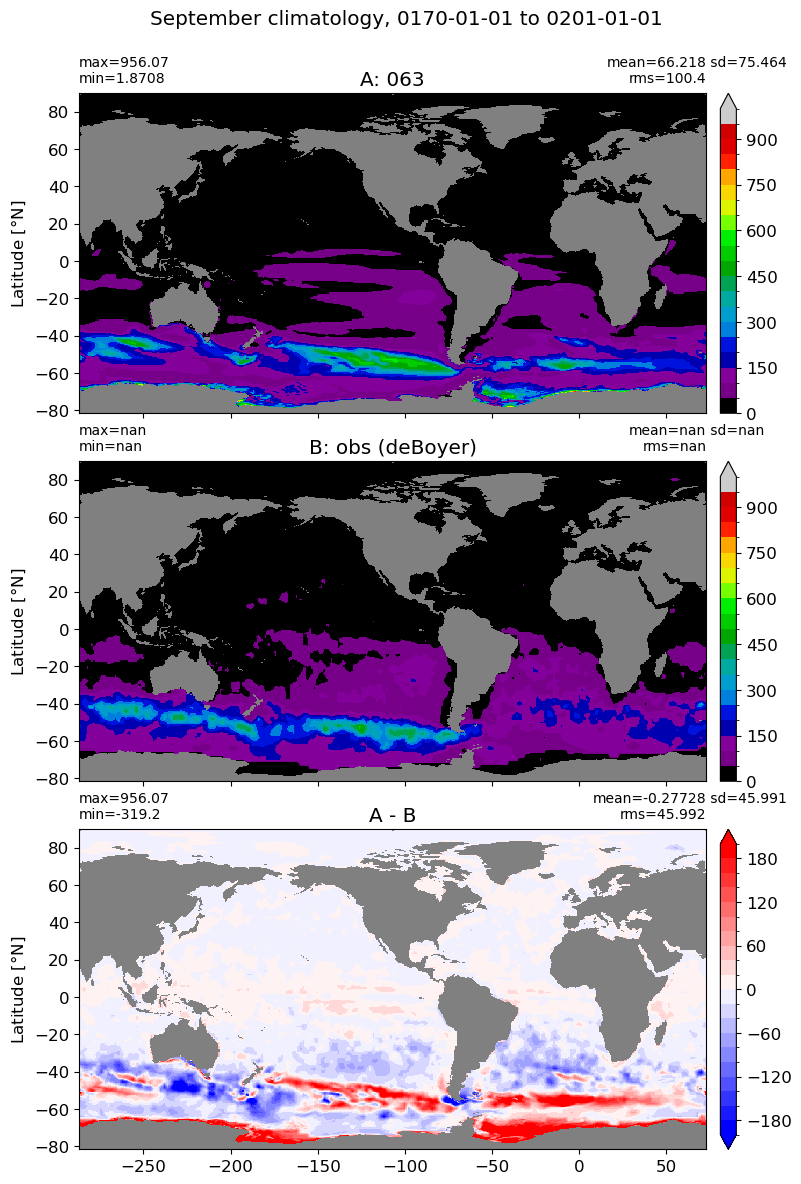
September#
m=8
for i in range(len(label)):
model = np.ma.masked_invalid(ds[i].mlotst.isel(month=m).values)
obs = np.ma.masked_where(grd[i].wet == 0, mld_obs.mld.isel(month=m).values)
xycompare(model,
obs,
grd[i].geolon, grd[i].geolat, grd[0].areacello,
title1 = label[i],
title2 = 'obs (deBoyer)',
suptitle=str(months[m]) +' climatology, '+ str(start_date) + ' to ' + str(end_date),
colormap=plt.cm.nipy_spectral, dcolormap=plt.cm.bwr,
clim = (0,1000), extend='max',
dlim=(-200,200))
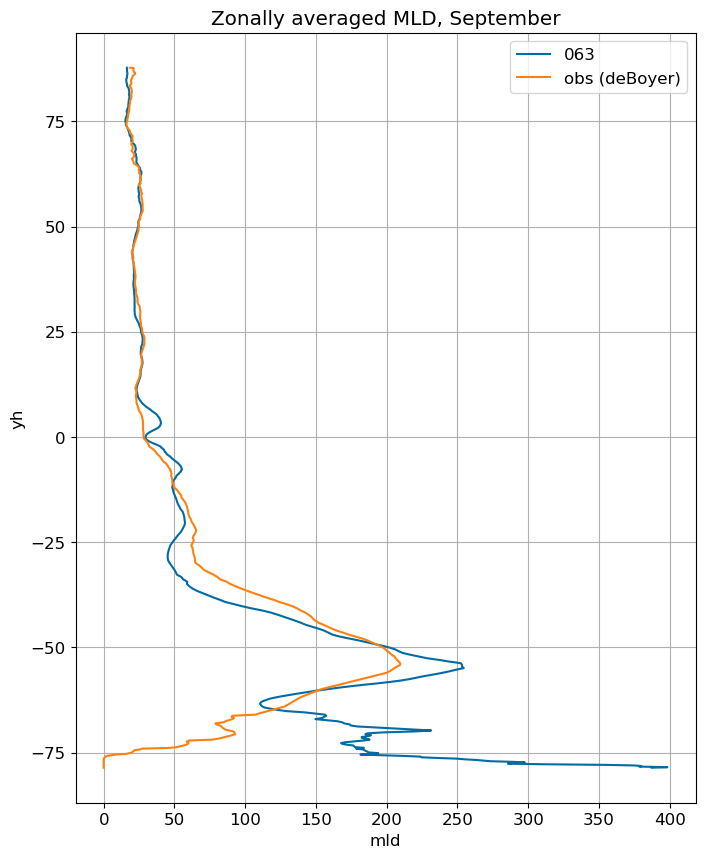
fig, ax = plt.subplots(figsize=(8,10))
for i in range(len(label)):
ds[i].mlotst.isel(month=m).weighted(grd_xr[i].areacello.fillna(0)).mean('xh').plot(y="yh",
ax=ax, label=label[i])
mld_obs.mld.isel(month=m).weighted(grd_xr[0].areacello.fillna(0)).mean('xh').plot(y="yh",
ax=ax, label='obs (deBoyer)')
ax.set_title('Zonally averaged MLD, '+str(months[m]))
ax.grid()
ax.legend();
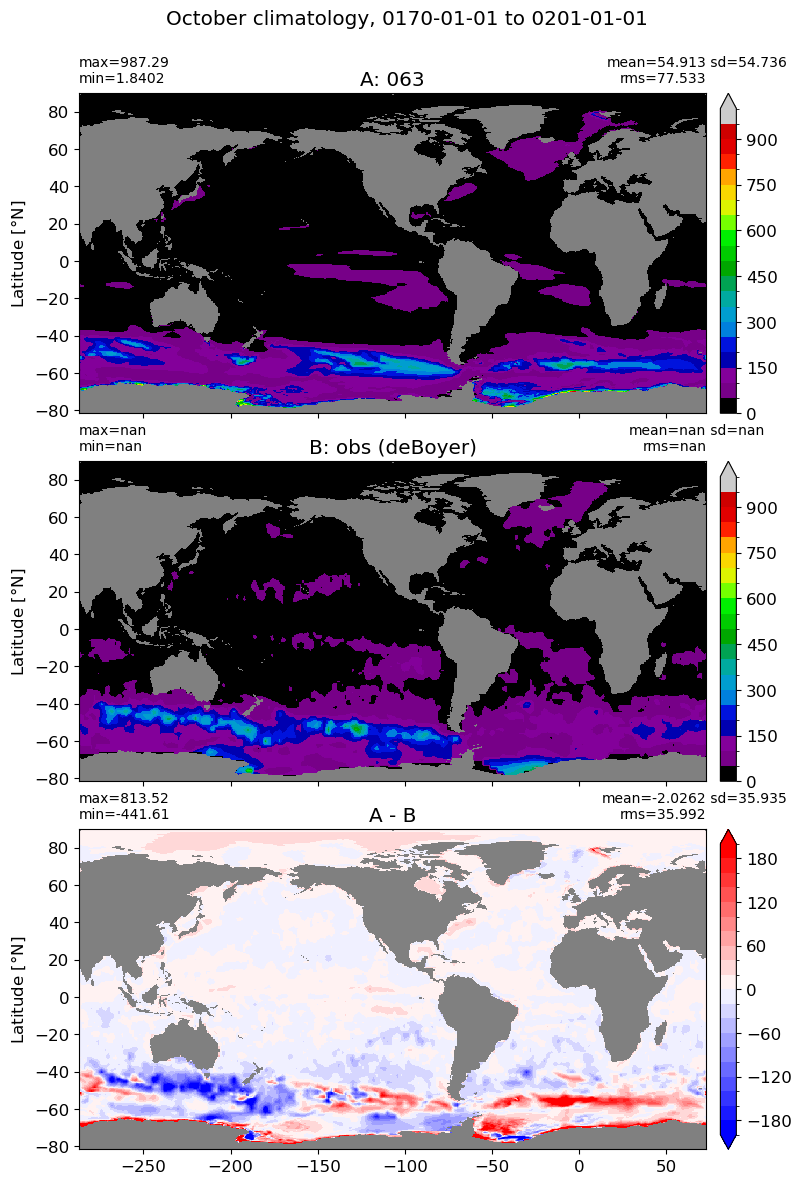
October#
m=9
for i in range(len(label)):
model = np.ma.masked_invalid(ds[i].mlotst.isel(month=m).values)
obs = np.ma.masked_where(grd[i].wet == 0, mld_obs.mld.isel(month=m).values)
xycompare(model,
obs,
grd[i].geolon, grd[i].geolat, grd[0].areacello,
title1 = label[i],
title2 = 'obs (deBoyer)',
suptitle=str(months[m]) +' climatology, '+ str(start_date) + ' to ' + str(end_date),
colormap=plt.cm.nipy_spectral, dcolormap=plt.cm.bwr,
clim = (0,1000), extend='max',
dlim=(-200,200))
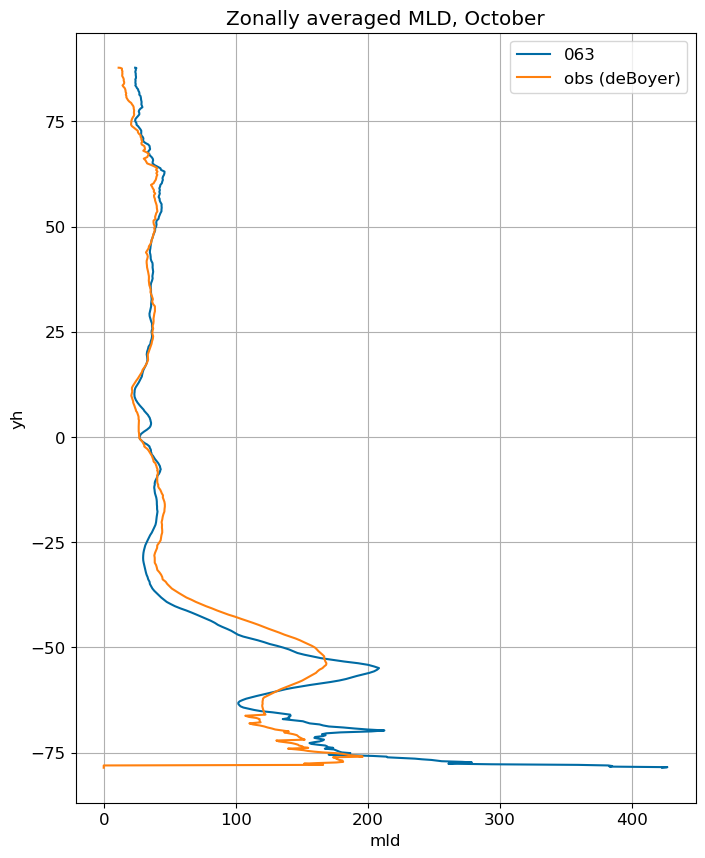
fig, ax = plt.subplots(figsize=(8,10))
for i in range(len(label)):
ds[i].mlotst.isel(month=m).weighted(grd_xr[i].areacello.fillna(0)).mean('xh').plot(y="yh",
ax=ax, label=label[i])
mld_obs.mld.isel(month=m).weighted(grd_xr[0].areacello.fillna(0)).mean('xh').plot(y="yh",
ax=ax, label='obs (deBoyer)')
ax.set_title('Zonally averaged MLD, '+str(months[m]))
ax.grid()
ax.legend();
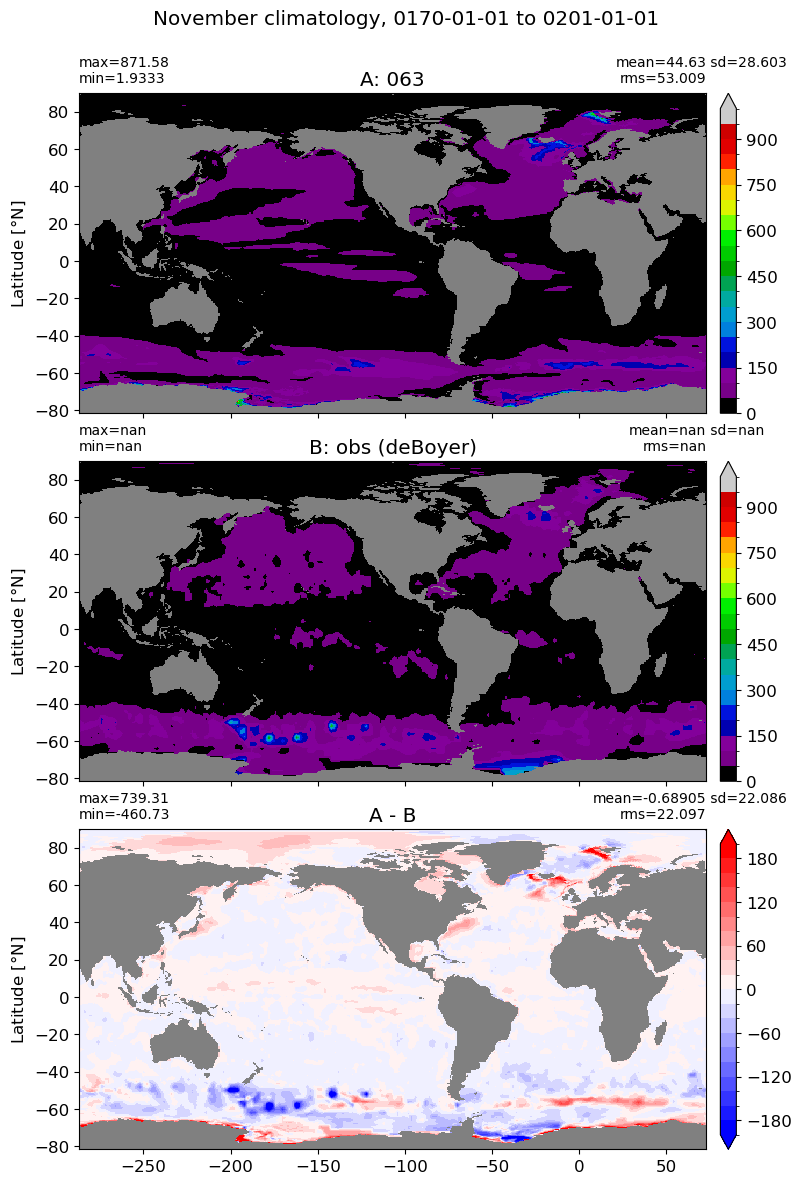
November#
m=10
for i in range(len(label)):
model = np.ma.masked_invalid(ds[i].mlotst.isel(month=m).values)
obs = np.ma.masked_where(grd[i].wet == 0, mld_obs.mld.isel(month=m).values)
xycompare(model,
obs,
grd[i].geolon, grd[i].geolat, grd[0].areacello,
title1 = label[i],
title2 = 'obs (deBoyer)',
suptitle=str(months[m]) +' climatology, '+ str(start_date) + ' to ' + str(end_date),
colormap=plt.cm.nipy_spectral, dcolormap=plt.cm.bwr,
clim = (0,1000), extend='max',
dlim=(-200,200))
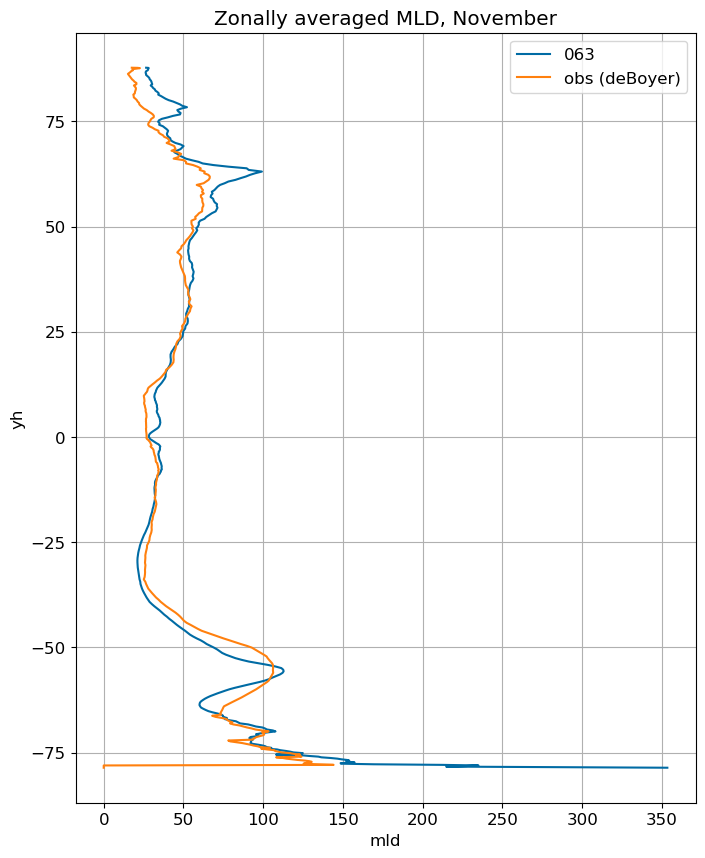
fig, ax = plt.subplots(figsize=(8,10))
for i in range(len(label)):
ds[i].mlotst.isel(month=m).weighted(grd_xr[i].areacello.fillna(0)).mean('xh').plot(y="yh",
ax=ax, label=label[i])
mld_obs.mld.isel(month=m).weighted(grd_xr[0].areacello.fillna(0)).mean('xh').plot(y="yh",
ax=ax, label='obs (deBoyer)')
ax.set_title('Zonally averaged MLD, '+str(months[m]))
ax.grid()
ax.legend();
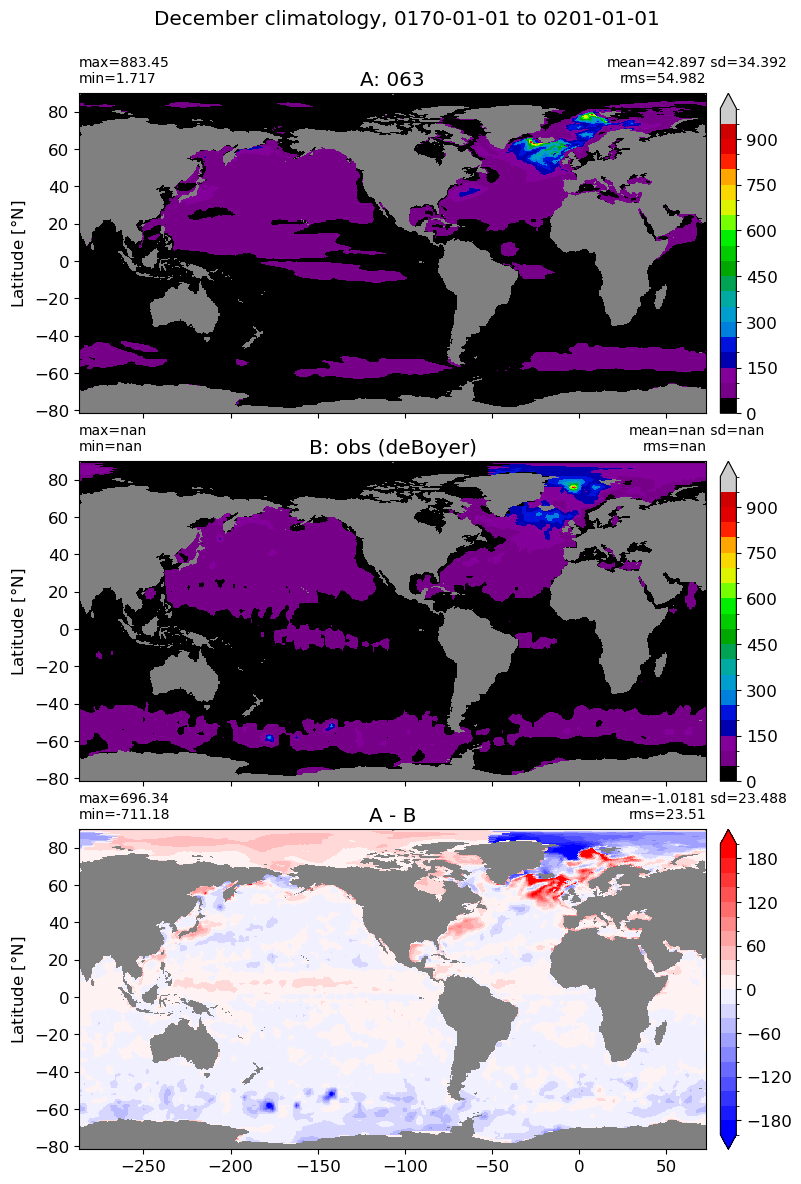
December#
m=11
for i in range(len(label)):
model = np.ma.masked_invalid(ds[i].mlotst.isel(month=m).values)
obs = np.ma.masked_where(grd[i].wet == 0, mld_obs.mld.isel(month=m).values)
xycompare(model,
obs,
grd[i].geolon, grd[i].geolat, grd[0].areacello,
title1 = label[i],
title2 = 'obs (deBoyer)',
suptitle=str(months[m]) +' climatology, '+ str(start_date) + ' to ' + str(end_date),
colormap=plt.cm.nipy_spectral, dcolormap=plt.cm.bwr,
clim = (0,1000), extend='max',
dlim=(-200,200))
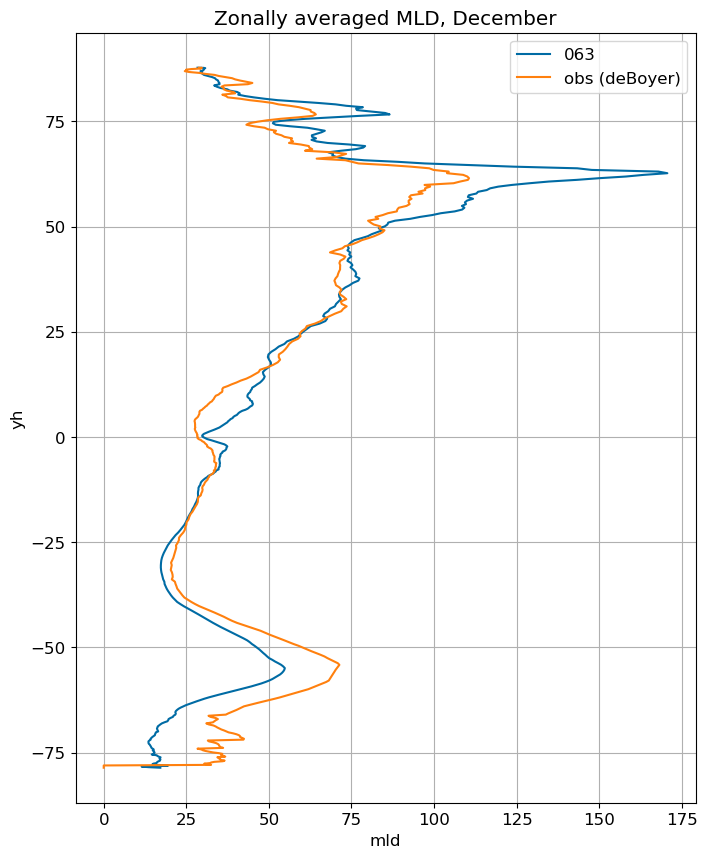
fig, ax = plt.subplots(figsize=(8,10))
for i in range(len(label)):
ds[i].mlotst.isel(month=m).weighted(grd_xr[i].areacello.fillna(0)).mean('xh').plot(y="yh",
ax=ax, label=label[i])
mld_obs.mld.isel(month=m).weighted(grd_xr[0].areacello.fillna(0)).mean('xh').plot(y="yh",
ax=ax, label='obs (deBoyer)')
ax.set_title('Zonally averaged MLD, '+str(months[m]))
ax.grid()
ax.legend();
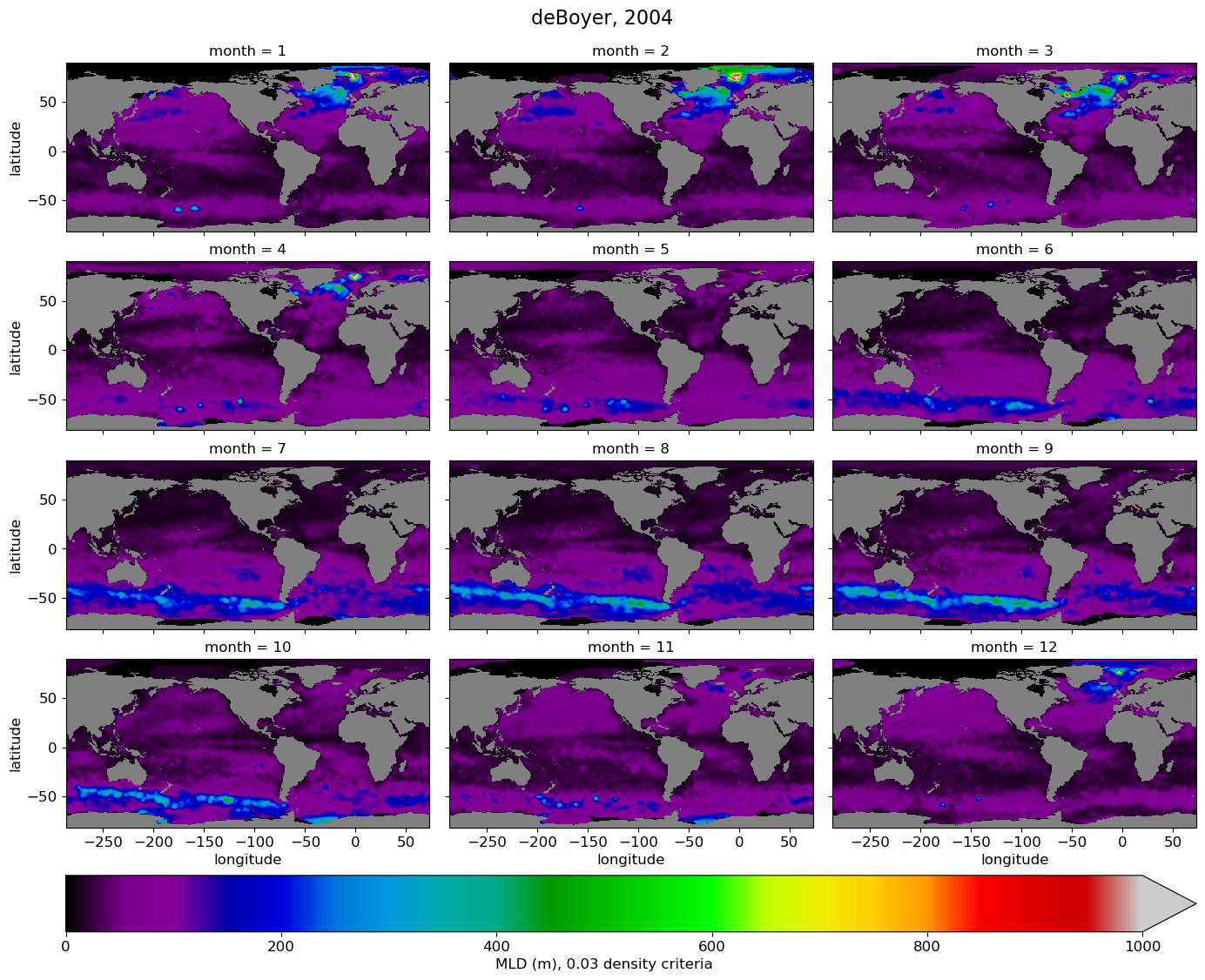
All months, obs#
cmap = plt.cm.nipy_spectral.copy()
cmap.set_bad(color='gray')
g = mld_obs.mld.plot(x="longitude", y="latitude", col='month',
col_wrap=3,
robust=True,
figsize=(14,12),
cmap=cmap,
vmin=0., vmax=1000.,
cbar_kwargs={"orientation": "horizontal", "pad": 0.05, "label": 'MLD (m), 0.03 density criteria'},
)
# Add a suptitle
g.fig.suptitle('deBoyer, 2004', fontsize=16, y=1.02);
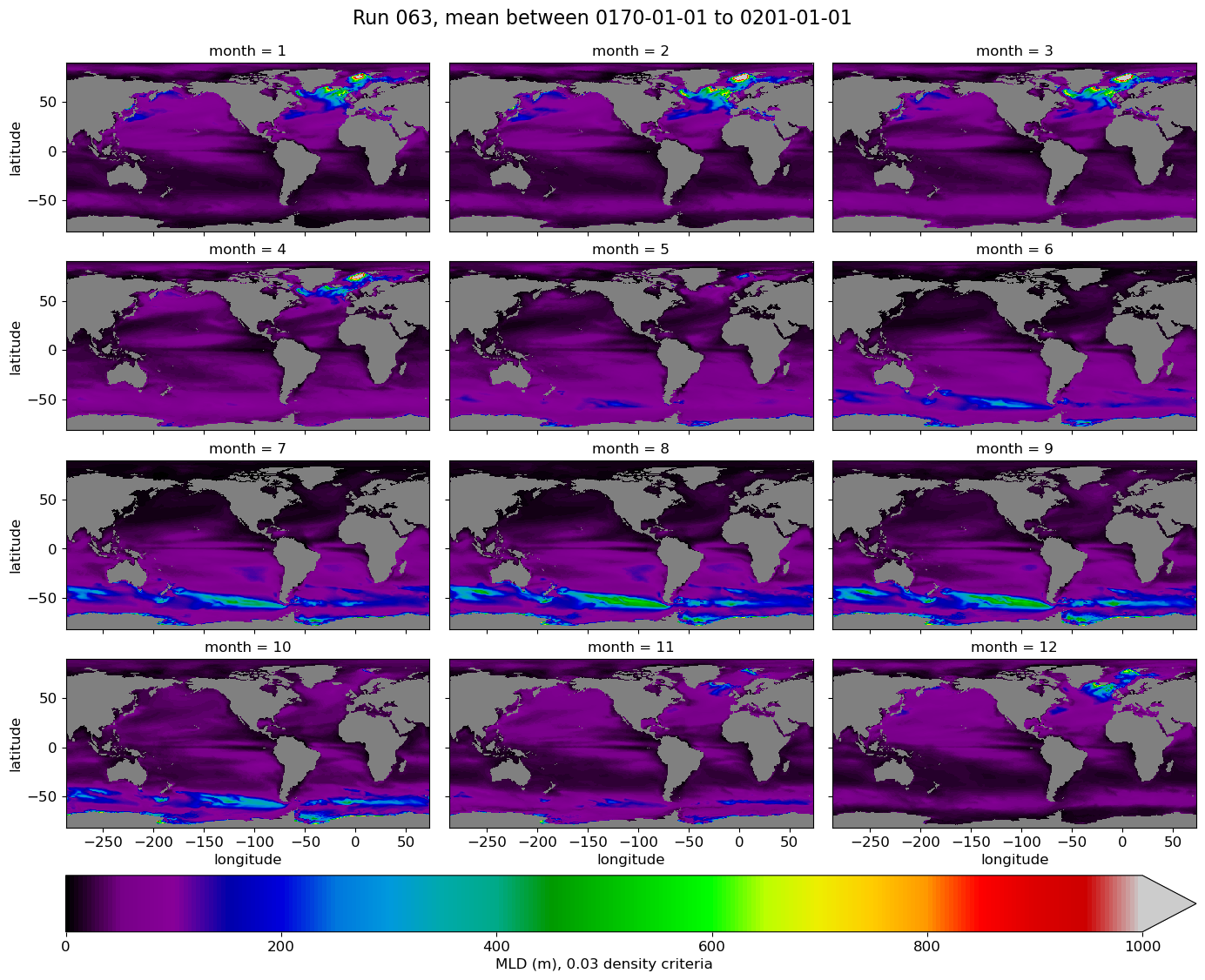
All months, model#
for path, case, i in zip(ocn_path, casename, range(len(casename))):
ds = xr.open_dataset(path+case+'_MLD_monthly_clima.nc')
g = ds.mlotst.plot(x="longitude", y="latitude", col='month',
col_wrap=3,
robust=True,
figsize=(14,12),
cmap=cmap,
vmin=0., vmax=1000.,
cbar_kwargs={"orientation": "horizontal", "pad": 0.05, "label": 'MLD (m), 0.03 density criteria'},
)
# Add a suptitle
g.fig.suptitle('Run {}, mean between {} to {}'.format(label[i], start_date, end_date), fontsize=16, y=1.02);
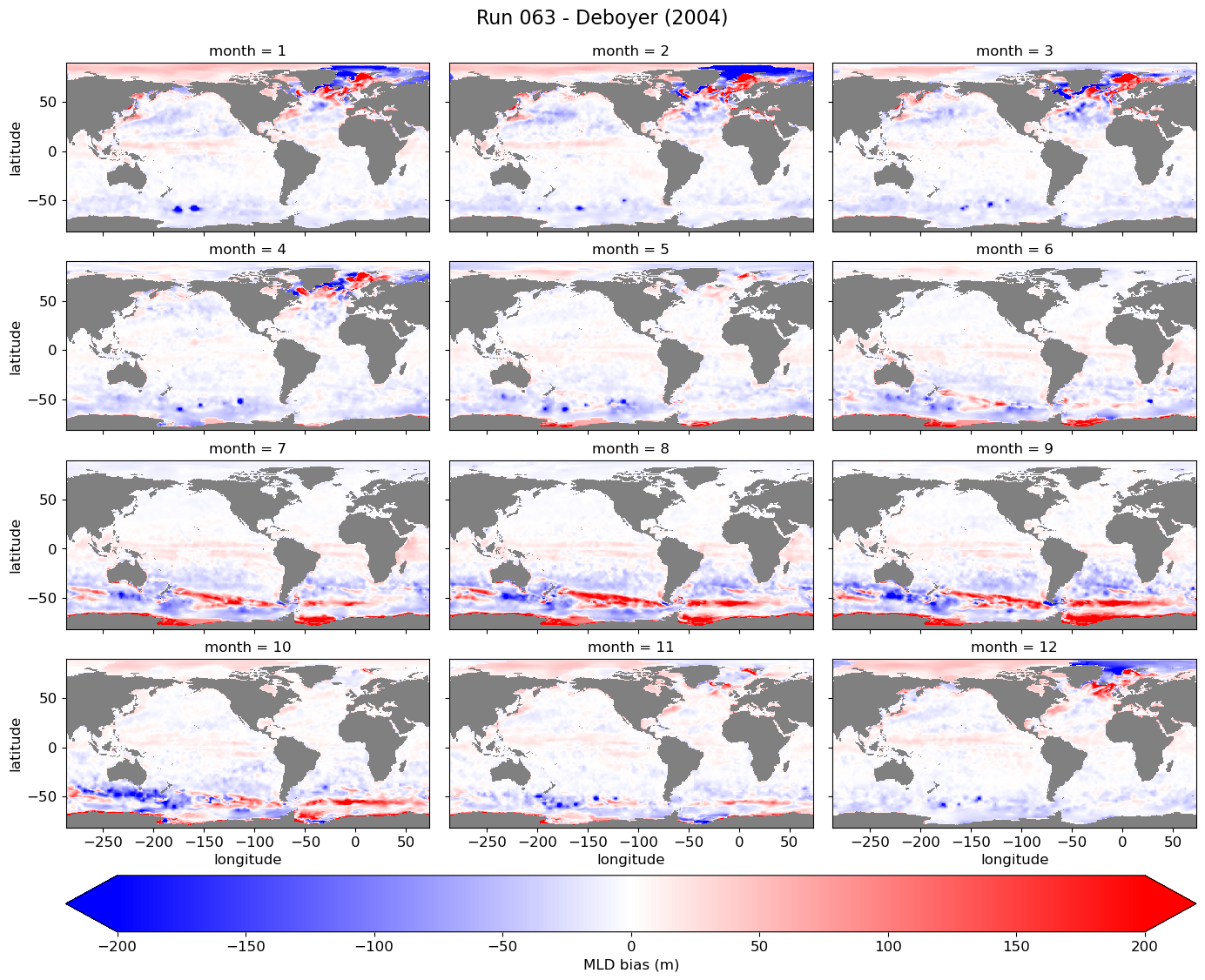
All months, model - obs#
cmap = plt.cm.bwr.copy()
cmap.set_bad(color='gray')
for path, case, i in zip(ocn_path, casename, range(len(casename))):
ds = xr.open_dataset(path+case+'_MLD_monthly_clima.nc')
diff = ds.mlotst - mld_obs.mld
g = diff.plot(x="longitude", y="latitude", col='month',
col_wrap=3,
robust=True,
figsize=(14,12),
cmap=cmap,
vmin=-200, vmax=200.,
cbar_kwargs={"orientation": "horizontal", "pad": 0.05, "label": 'MLD bias (m)'},
)
# Add a suptitle
g.fig.suptitle('Run {} - Deboyer (2004)'.format(label[i]), fontsize=16, y=1.02);
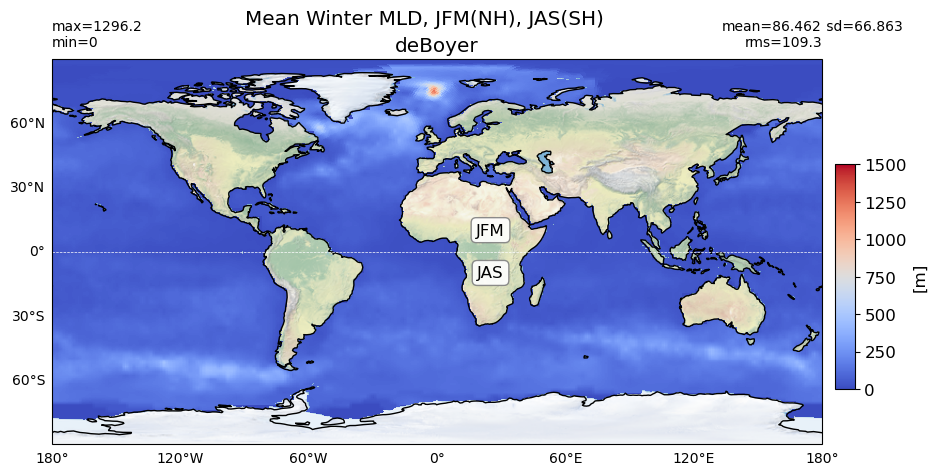
Winter#
%matplotlib inline
title = 'Mean Winter MLD, JFM(NH), JAS(SH)'
try:
area= grd[0].area_t
except:
area= grd[0].areacello
plot_map(obs_winter, area, grd[0], 'deBoyer',
vmin=0, vmax=1500, suptitle=title)
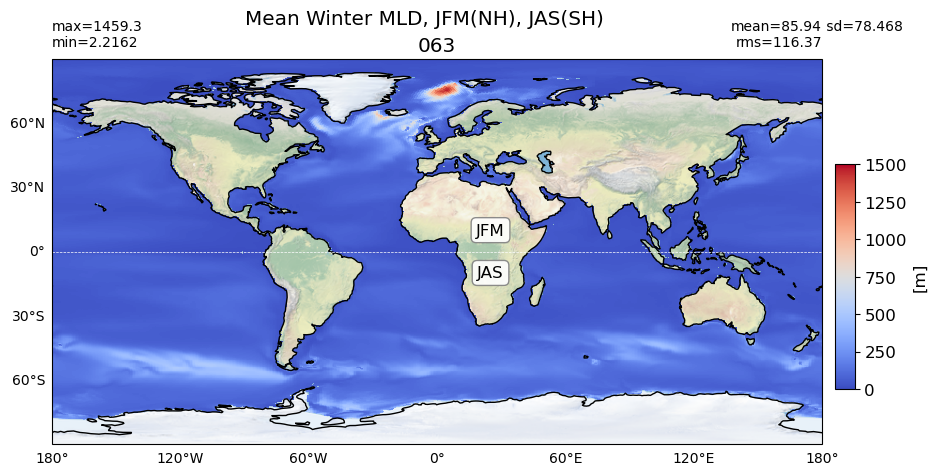
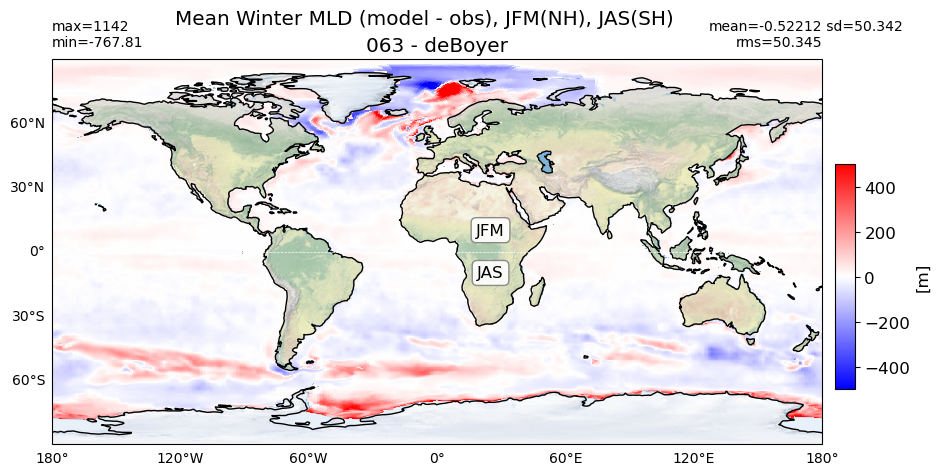
title1 = 'Mean Winter MLD, JFM(NH), JAS(SH)'
title2 = 'Mean Winter MLD (model - obs), JFM(NH), JAS(SH)'
for path, case, i in zip(ocn_path, casename, range(len(casename))):
da = xr.open_dataset(path+case+'_MLD_winter.nc')
try:
area= grd[i].area_t
except:
area= grd[i].areacello
# model
plot_map(da.MLD_winter.values, area, grd[i], label[i],
vmin=0, vmax=1500, suptitle=title1)
# model - obs
diff = (da.MLD_winter.values - obs_winter)
plot_map(diff, area, grd[i], label[i] + ' - deBoyer',
vmin=-500, vmax=500, suptitle=title2, cmap='bwr')
obs_winter
masked_array(
data=[[--, --, --, ..., --, --, --],
[--, --, --, ..., --, --, --],
[--, --, --, ..., --, --, --],
...,
[--, --, --, ..., --, --, --],
[--, --, --, ..., --, --, --],
[--, --, --, ..., --, --, --]],
mask=[[ True, True, True, ..., True, True, True],
[ True, True, True, ..., True, True, True],
[ True, True, True, ..., True, True, True],
...,
[ True, True, True, ..., True, True, True],
[ True, True, True, ..., True, True, True],
[ True, True, True, ..., True, True, True]],
fill_value=1e+20,
dtype=float32)
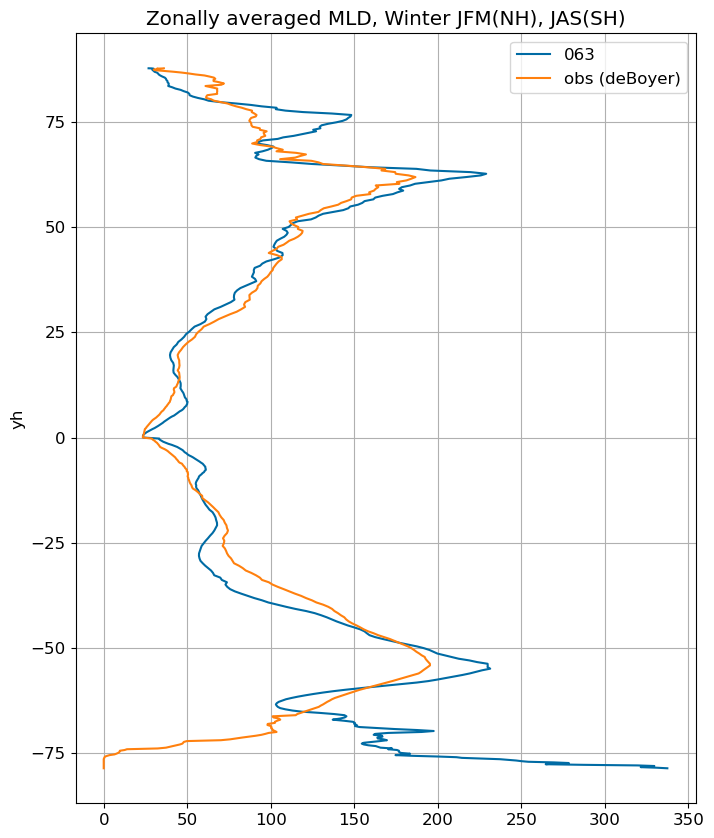
fig, ax = plt.subplots(figsize=(8,10))
for i in range(len(label)):
da = xr.open_dataset(path+case+'_MLD_winter.nc')
da.MLD_winter.weighted(grd_xr[i].areacello.fillna(0)).mean('xh').plot(y="yh",
ax=ax, label=label[i])
obs_winter_da = xr.DataArray(
data=obs_winter, # New data (all zeros)
coords=da.coords,
dims=da.dims
)
obs_winter_da.weighted(grd_xr[0].areacello.fillna(0)).mean('xh').plot(y="yh",
ax=ax, label='obs (deBoyer)')
ax.set_title('Zonally averaged MLD, Winter JFM(NH), JAS(SH)')
ax.grid()
ax.legend();
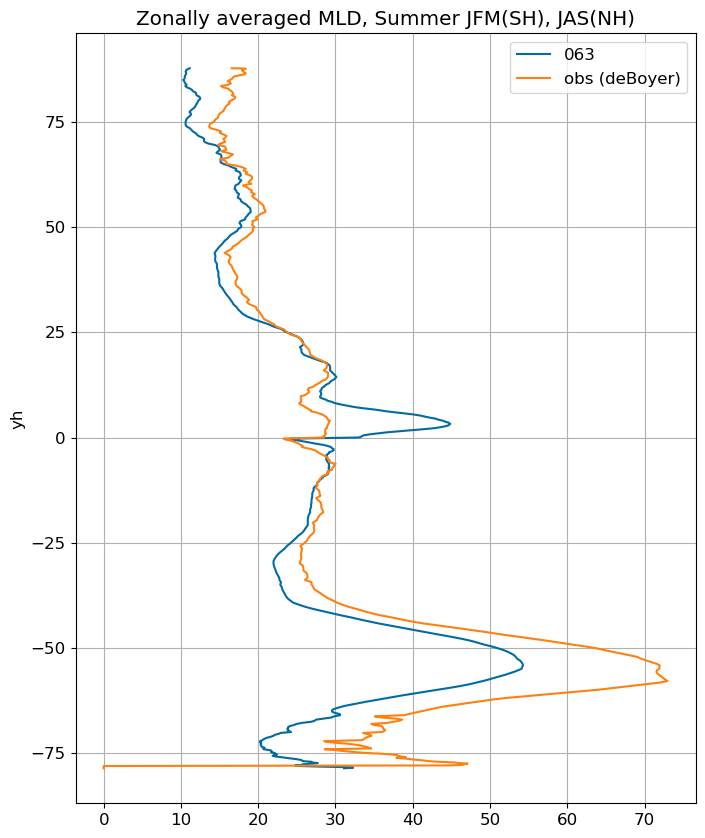
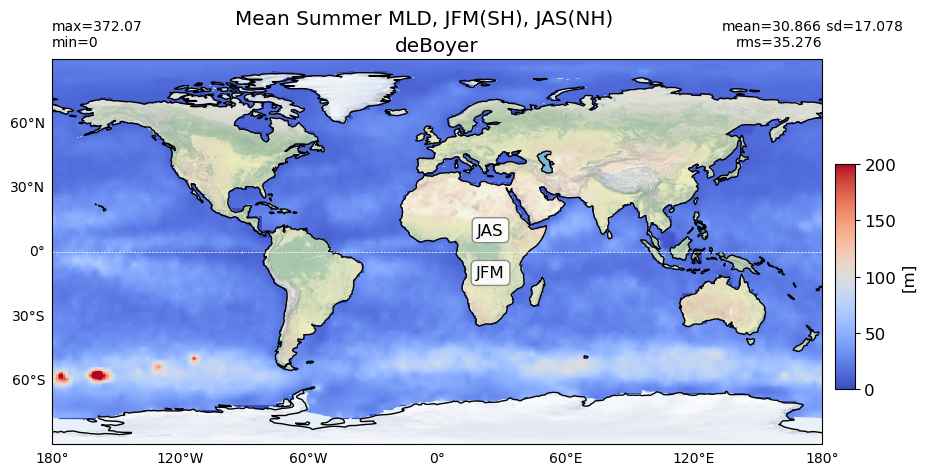
Summer#
title = 'Mean Summer MLD, JFM(SH), JAS(NH)'
try:
area= grd[0].area_t
except:
area= grd[0].areacello
plot_map(obs_summer, area, grd[0], 'deBoyer', vmin=0, vmax=200,
suptitle=title, nh='JAS', sh='JFM',)
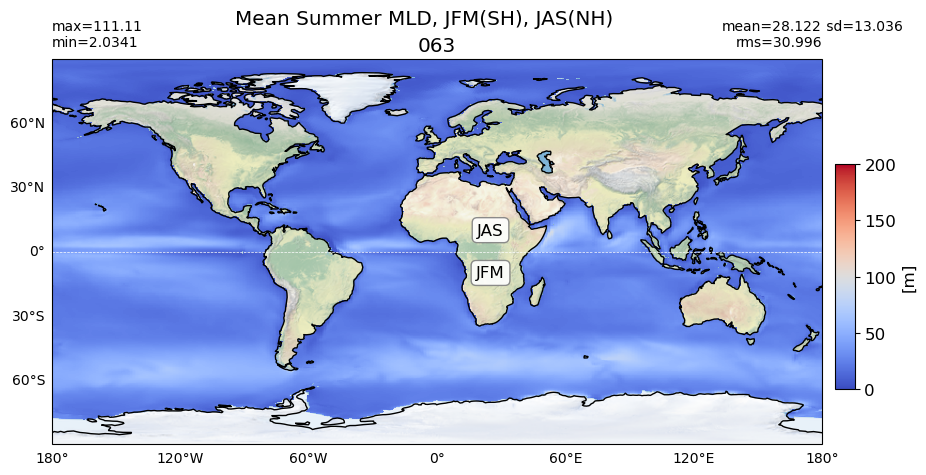
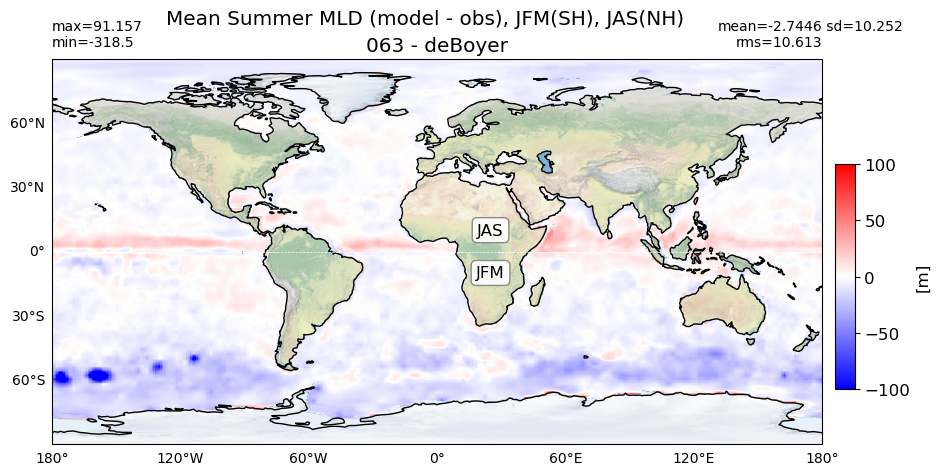
title1 = 'Mean Summer MLD, JFM(SH), JAS(NH)'
title2 = 'Mean Summer MLD (model - obs), JFM(SH), JAS(NH)'
for path, case, i in zip(ocn_path, casename, range(len(casename))):
da = xr.open_dataset(path+case+'_MLD_summer.nc')
try:
area= grd[i].area_t
except:
area= grd[i].areacello
# model
plot_map(da.MLD_summer.values, area, grd[i], label[i],
vmin=0, vmax=200, suptitle=title1, nh='JAS', sh='JFM')
# model - obs
diff = (da.MLD_summer.values - obs_summer)
plot_map(diff, area, grd[i], label[i] + ' - deBoyer',
vmin=-100, vmax=100, suptitle=title2, nh='JAS',
sh='JFM',cmap='bwr')
fig, ax = plt.subplots(figsize=(8,10))
for i in range(len(label)):
da = xr.open_dataset(path+case+'_MLD_summer.nc')
da.MLD_summer.weighted(grd_xr[i].areacello.fillna(0)).mean('xh').plot(y="yh",
ax=ax, label=label[i])
obs_summer_da = xr.DataArray(
data=obs_summer, # New data (all zeros)
coords=da.coords,
dims=da.dims
)
obs_summer_da.weighted(grd_xr[0].areacello.fillna(0)).mean('xh').plot(y="yh",
ax=ax, label='obs (deBoyer)')
ax.set_title('Zonally averaged MLD, Summer JFM(SH), JAS(NH)')
ax.grid()
ax.legend();