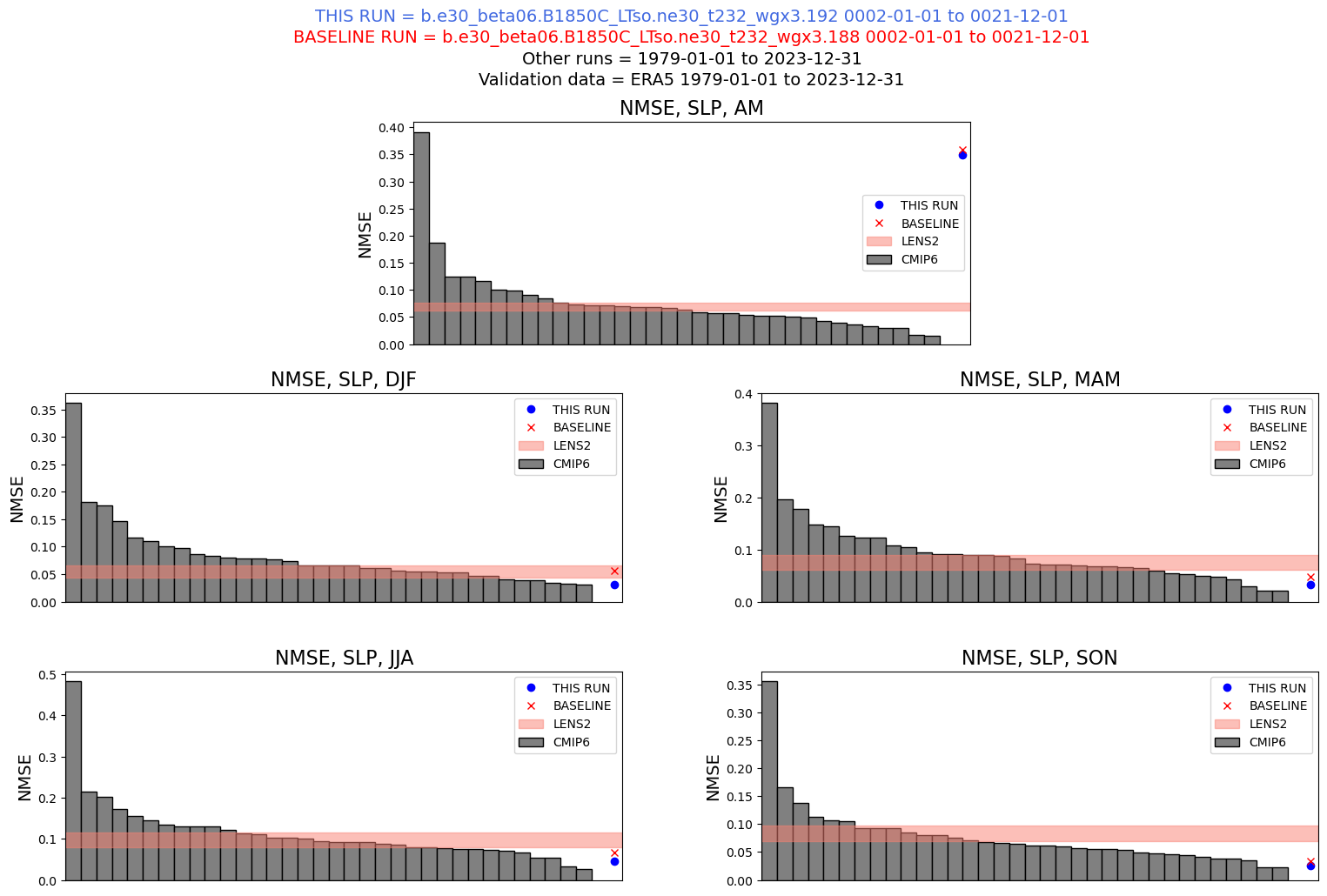
Normalized Mean Square Error#
This notebook computes the normalized mean square error of atmospheric surface pressure. It is compared to ERA5 observations, as well as the CESM2 large ensemble and CMIP6 model output.
Parameters#
These variables are set in config.yml
# Parameters
case_name = "b.e30_beta06.B1850C_LTso.ne30_t232_wgx3.192"
base_case_name = "b.e30_beta06.B1850C_LTso.ne30_t232_wgx3.188"
CESM_output_dir = "/glade/derecho/scratch/hannay/archive"
base_case_output_dir = "/glade/derecho/scratch/gmarques/archive"
start_date = "0002-01-01"
end_date = "0021-12-01"
base_start_date = "0002-01-01"
base_end_date = "0021-12-01"
obs_data_dir = (
"/glade/campaign/cesm/development/cross-wg/diagnostic_framework/CUPiD_obs_data"
)
ts_dir = None
lc_kwargs = {"threads_per_worker": 1}
serial = True
regridded_output = False
base_regridded_output = False
validation_path = (
"atm/analysis_datasets/fv0.9x1.25/seasonal_climatology/nmse_validation/PSL/"
)
subset_kwargs = {}
product = "/glade/work/hannay/CUPiD/examples/key_metrics/computed_notebooks//atm/Global_PSL_NMSE_compare_obs_lens.ipynb"
Read in the current case#
if regridded_output:
file_path = f"{ts_dir}/{case_name}/atm/proc/tseries/regrid"
else:
file_path = f"{ts_dir}/{case_name}/atm/proc/tseries"
print(file_path)
/glade/derecho/scratch/hannay/archive/b.e30_beta06.B1850C_LTso.ne30_t232_wgx3.192/atm/proc/tseries
if base_case_name is not None:
if base_regridded_output:
base_file_path = f"{ts_dir}/{base_case_name}/atm/proc/tseries/regrid"
else:
base_file_path = f"{ts_dir}/{base_case_name}/atm/proc/tseries"
print(base_file_path)
/glade/derecho/scratch/hannay/archive/b.e30_beta06.B1850C_LTso.ne30_t232_wgx3.188/atm/proc/tseries
Read in validation data and other CMIP models for comparison (precomputed)#
Compute the NMSE#
Set up the plot panel#