Land ice SMB model comparison#
This notebook compares the downscaled output of surface mass balance (SMB) over the Greenland ice sheet (GrIS) to the regional model MAR. In what follows, we interchangeably call the MAR data “observation”.
Note1: the MAR data are processed as a climatology spanning 1960-1999.
Note2: the MAR data are available at a uniform resolution of 1km using the same projection as the CISM grid. This notebook requires the interpolation of the MAR data on the CISM grid. The interpolation is done in this notebook (for now) to allow for the eventuality of the CISM grid or the MAR grid to change in the future.
creation: 05-26-24
contact: Gunter Leguy (gunterl@ucar.edu)
Parameter configuration#
Some parameters are set in CUPiD’s config.yml file,
others are derived from these parameters.
# Parameters
case_name = "b.e30_beta06.B1850C_LTso.ne30_t232_wgx3.192"
base_case_name = "b.e30_beta06.B1850C_LTso.ne30_t232_wgx3.188"
CESM_output_dir = "/glade/derecho/scratch/hannay/archive"
base_case_output_dir = "/glade/derecho/scratch/gmarques/archive"
start_date = "0002-01-01"
end_date = "0021-12-01"
base_start_date = "0002-01-01"
base_end_date = "0021-12-01"
obs_data_dir = (
"/glade/campaign/cesm/development/cross-wg/diagnostic_framework/CUPiD_obs_data"
)
ts_dir = None
lc_kwargs = {"threads_per_worker": 1}
serial = True
obs_path = "glc/analysis_datasets/multi_grid/annual_avg/SMB_data"
obs_name = "GrIS_MARv3.12_climo_1960_1999.nc"
climo_nyears = 40
subset_kwargs = {}
product = "/glade/work/hannay/CUPiD/examples/key_metrics/computed_notebooks//glc/Greenland_SMB_visual_compare_obs.ipynb"
Set up grid#
Read in the grid data, compute resolution and other grid-specific parameters
Make datasets#
Read in observations and CESM output. Also do necessary computations (global mean for time series, temporal mean for climatology).
Generate plots#
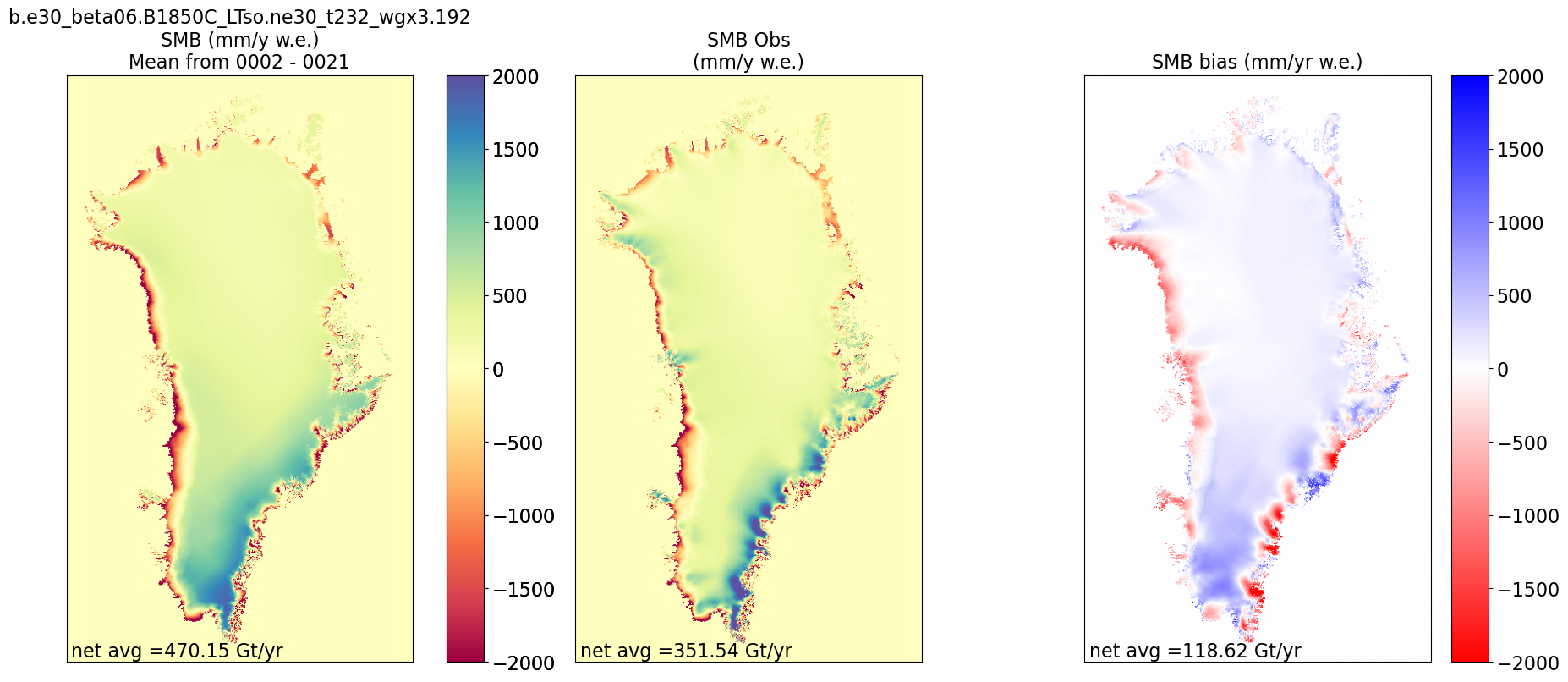
Map comparing CESM to observation, possibly map comparing CESM to older case, and time series of spatial mean SMB.
/glade/derecho/scratch/hannay/tmp/ipykernel_96314/1639303292.py:4: MatplotlibDeprecationWarning: The get_cmap function was deprecated in Matplotlib 3.7 and will be removed in 3.11. Use ``matplotlib.colormaps[name]`` or ``matplotlib.colormaps.get_cmap()`` or ``pyplot.get_cmap()`` instead.
my_cmap = mcm.get_cmap("Spectral")
/glade/derecho/scratch/hannay/tmp/ipykernel_96314/1639303292.py:5: MatplotlibDeprecationWarning: The get_cmap function was deprecated in Matplotlib 3.7 and will be removed in 3.11. Use ``matplotlib.colormaps[name]`` or ``matplotlib.colormaps.get_cmap()`` or ``pyplot.get_cmap()`` instead.
my_cmap_diff = mcm.get_cmap("bwr_r")
/glade/derecho/scratch/hannay/tmp/ipykernel_96314/2450413827.py:4: MatplotlibDeprecationWarning: The get_cmap function was deprecated in Matplotlib 3.7 and will be removed in 3.11. Use ``matplotlib.colormaps[name]`` or ``matplotlib.colormaps.get_cmap()`` or ``pyplot.get_cmap()`` instead.
my_cmap = mcm.get_cmap("Spectral")
/glade/derecho/scratch/hannay/tmp/ipykernel_96314/2450413827.py:5: MatplotlibDeprecationWarning: The get_cmap function was deprecated in Matplotlib 3.7 and will be removed in 3.11. Use ``matplotlib.colormaps[name]`` or ``matplotlib.colormaps.get_cmap()`` or ``pyplot.get_cmap()`` instead.
my_cmap_diff = mcm.get_cmap("bwr_r")


