Meridional Overturning Circulation#
%load_ext autoreload
%autoreload 2
%%capture
# comment above line to see details about the run(s) displayed
from misc import *
from mom6_tools import m6plot, m6toolbox
from mom6_tools.moc import *
import glob
from IPython.display import Image, display
%matplotlib inline
ds = []
for c, l, p in zip(casename,label, ocn_path):
dummy = xr.open_dataset(p+'{}_MOC.nc'.format(c))
ds.append(dummy)
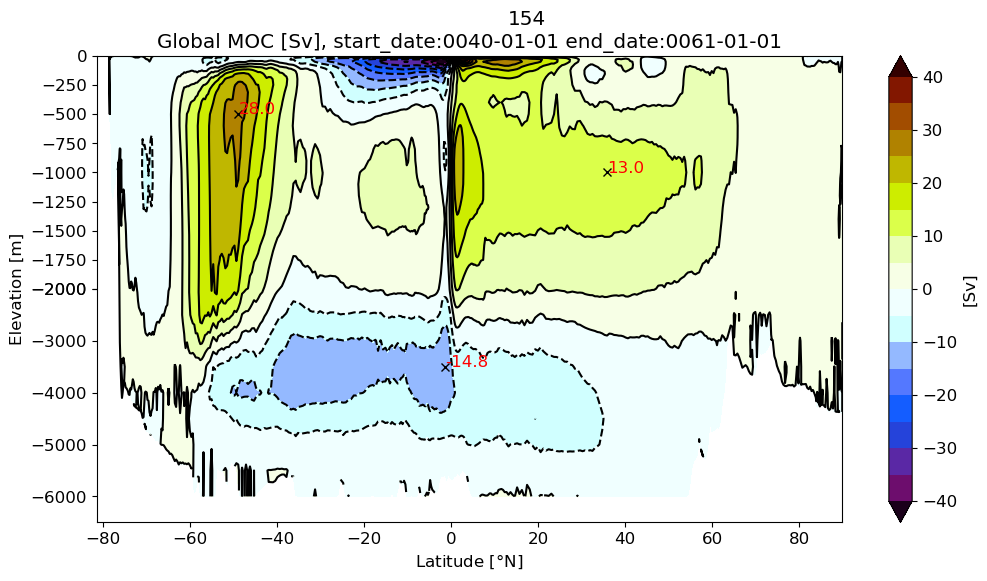
Global MOC#
pnum = len(ds)
varName = 'vmo'
Zmod = []
fname ='*.mom6.h.z.????-??.nc'
for i in range(pnum):
# this hack needs to be fixed
print(OUTDIR[i]+fname)
file = sorted(glob.glob(OUTDIR[i]+fname))[0:2]
ds1 = xr.open_mfdataset(file)
Zmod.append(m6toolbox.get_z(ds1, depth[i], varName))
/glade/derecho/scratch/gmarques/archive/g.e30_a07g.GW_JRA.TL319_t232_wgx3_hycom1_N75.2025.154/ocn/hist/*.mom6.h.z.????-??.nc
z#
for i in range(pnum):
m6plot.setFigureSize([16,9],576,debug=False)
axis = plt.gca()
cmap = plt.get_cmap('dunnePM')
zg = Zmod[i].min(axis=-1);
psiPlot = ds[i].moc.values
yyg = grd[i].geolat_c[:,:].max(axis=-1)+0*zg
ci=m6plot.pmCI(0.,40.,5.)
plotPsi(yyg, zg, psiPlot, ci, 'Global MOC [Sv], ' + \
'start_date:' + ds[i].start_date +' end_date:' + ds[i].end_date)
plt.xlabel(r'Latitude [$\degree$N]')
plt.suptitle(label[i])
findExtrema(yyg, zg, psiPlot, max_lat=-30.)
findExtrema(yyg, zg, psiPlot, min_lat=25., min_depth=250.)
findExtrema(yyg, zg, psiPlot, min_depth=2000., mult=-1.)
plt.gca().invert_yaxis()
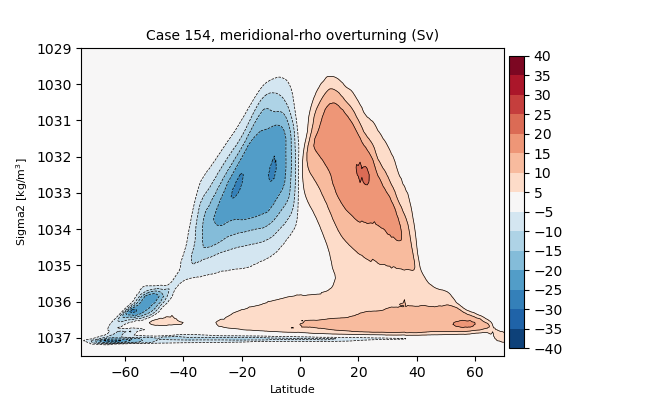
sigma2#
for i in range(len(casename)):
png_files = glob.glob(ocn_path[i]+'../PNG/MOC/'+casename[i]+'_MOC_sigma2_global.png')
for png_file in png_files:
display(Image(filename=png_file))
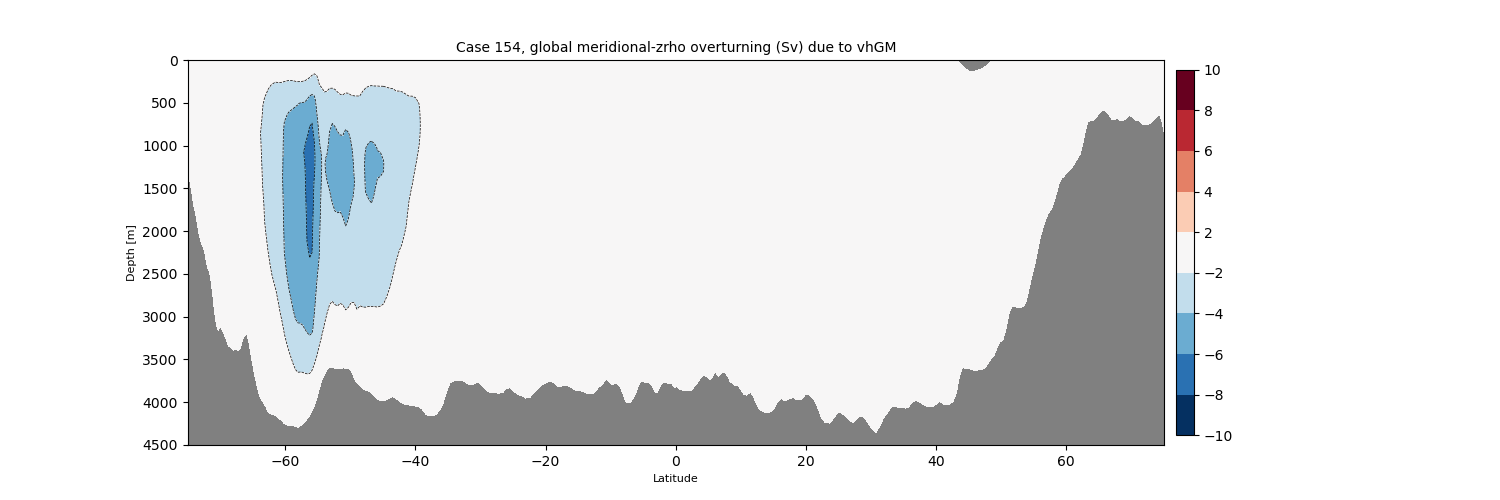
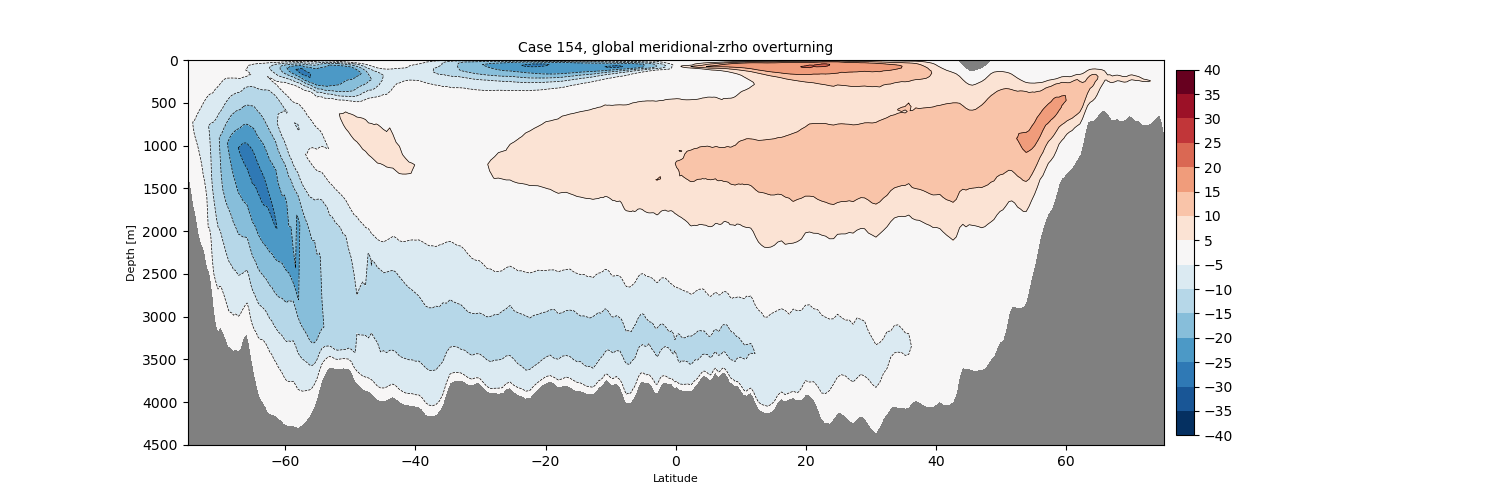
zrho#
for i in range(len(casename)):
png_files = glob.glob(ocn_path[i]+'../PNG/MOC/'+casename[i]+'_MOC_zrho_global.png')
for png_file in png_files:
display(Image(filename=png_file))
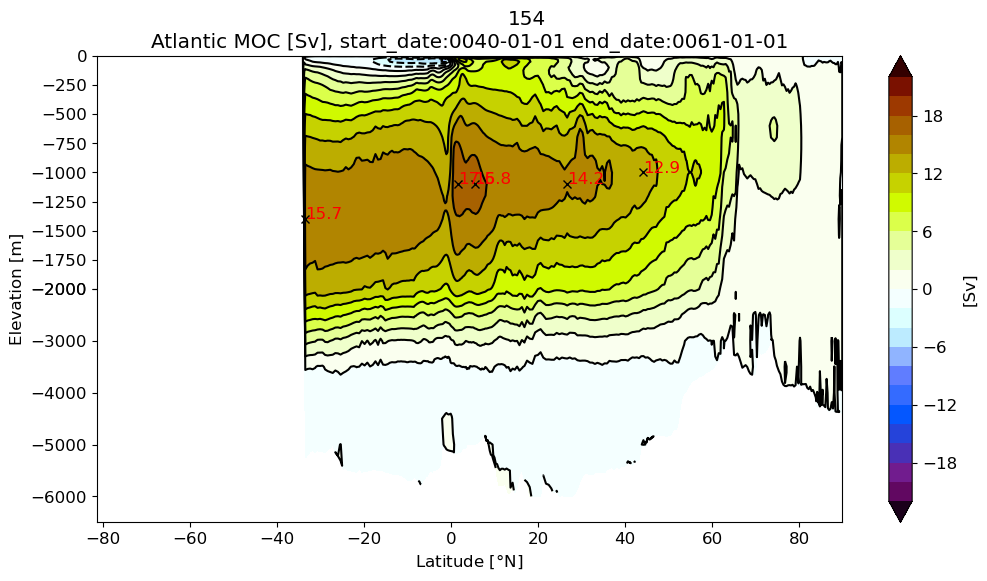
Atlantic MOC#
z#
# 2 - Atlatic; 4 - Arctic; 6 - Med; 7 - Baltic; 8 - Hudson Bay;
for i in range(pnum):
basin_code = genBasinMasks(grd[i].geolon, grd[i].geolat, depth[i], xda=False);
m6plot.setFigureSize([16,9],576,debug=False)
cmap = plt.get_cmap('dunnePM')
ci=m6plot.pmCI(0.,22.,2.)
m = 0*basin_code; m[(basin_code==2) | (basin_code==4) | (basin_code==6) | (basin_code==7) | (basin_code==8)]=1
z = (m*Zmod[i]).min(axis=-1)
psiPlot = ds[i].amoc.values
yy = grd[i].geolat_c[:,:].max(axis=-1)+0*z
plotPsi(yy, z, psiPlot, ci, 'Atlantic MOC [Sv], ' + \
'start_date:' + ds[i].start_date +' end_date:' + ds[i].end_date)
plt.xlabel(r'Latitude [$\degree$N]')
plt.suptitle(label[i])
findExtrema(yy, z, psiPlot, min_lat=26.5, max_lat=27., min_depth=250.) # RAPID
findExtrema(yy, z, psiPlot, min_lat=44, max_lat=46., min_depth=250.) # RAPID
findExtrema(yy, z, psiPlot, max_lat=-33.)
findExtrema(yy, z, psiPlot)
findExtrema(yy, z, psiPlot, min_lat=5.)
plt.gca().invert_yaxis()
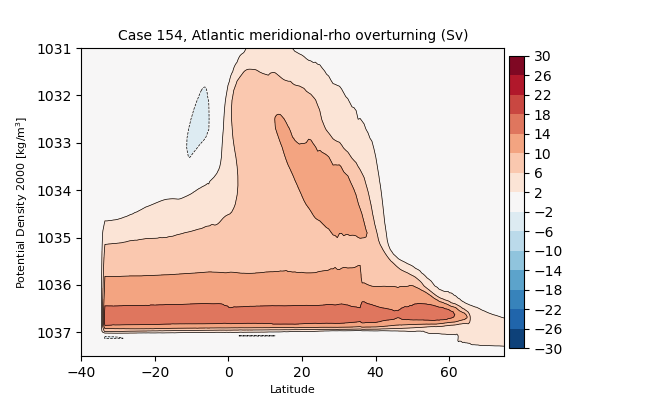
sigma2#
for i in range(len(casename)):
png_files = glob.glob(ocn_path[i]+'../PNG/MOC/'+casename[i]+'_MOC_sigma2_Atlantic.png')
for png_file in png_files:
display(Image(filename=png_file))
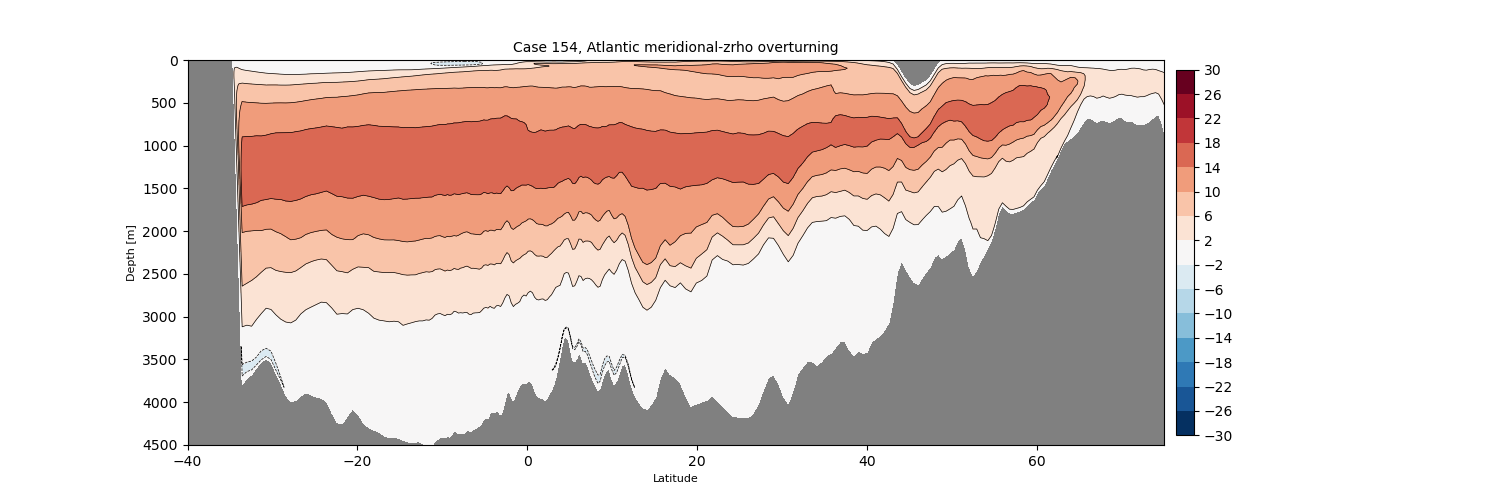
zrho#
for i in range(len(casename)):
png_files = glob.glob(ocn_path[i]+'../PNG/MOC/'+casename[i]+'_MOC_zrho_Atlantic.png')
for png_file in png_files:
display(Image(filename=png_file))
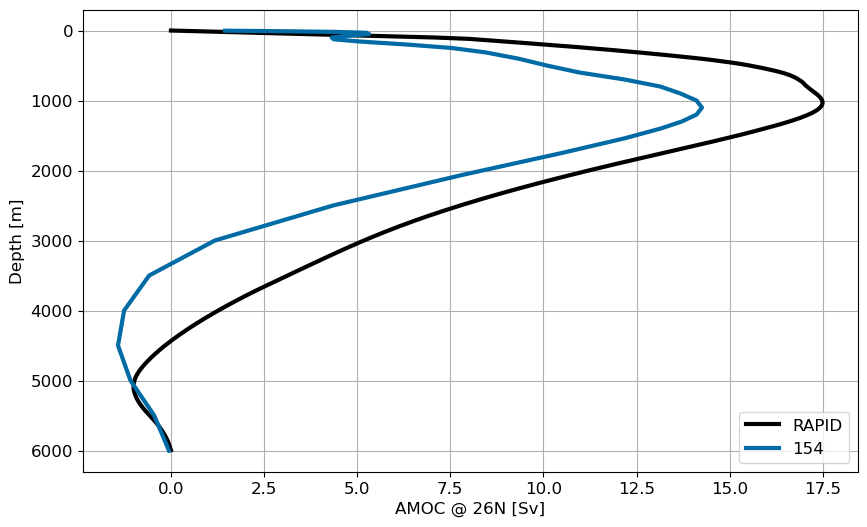
AMOC profile at 26N#
fig, ax = plt.subplots(figsize=(10,6))
rapid_vertical = xr.open_dataset('/glade/work/gmarques/cesm/datasets/RAPID/moc_vertical.nc')
ax.plot(rapid_vertical.stream_function_mar.mean('time'),
rapid_vertical.depth, 'k', label='RAPID', lw=3)
for i in range(pnum):
ax.plot(ds[i]['amoc'].sel(yq=26, method='nearest'), ds[i].zl, label=label[i], lw=3)
ax.legend()
plt.gca().invert_yaxis()
plt.grid()
ax.set_xlabel('AMOC @ 26N [Sv]')
ax.set_ylabel('Depth [m]');
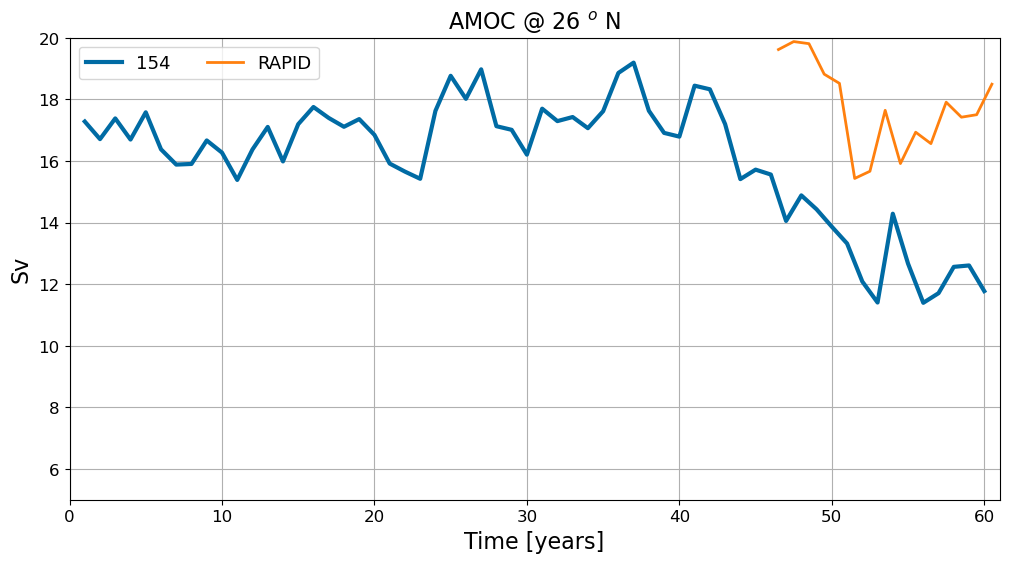
AMOC time series#
# load RAPID time series
rapid = m6toolbox.weighted_temporal_mean_vars(xr.open_dataset('/glade/work/gmarques/cesm/datasets/RAPID/moc_transports.nc'))
# plot
fig = plt.figure(figsize=(12, 6))
for i in range(pnum):
plt.plot(np.arange(len(ds[i].time))+1. ,ds[i]['amoc_26'].values,
label=label[i], lw=3)
# rapid
plt.plot(np.arange(len(rapid.time))+46.5 ,rapid.moc_mar_hc10.values,
label='RAPID', lw=2)
plt.title('AMOC @ 26 $^o$ N', fontsize=16)
plt.ylim(5,20)
plt.xlim(0,1+len(ds[0].time))
plt.xlabel('Time [years]', fontsize=16); plt.ylabel('Sv', fontsize=16)
plt.legend(fontsize=13, ncol=2)
plt.grid()
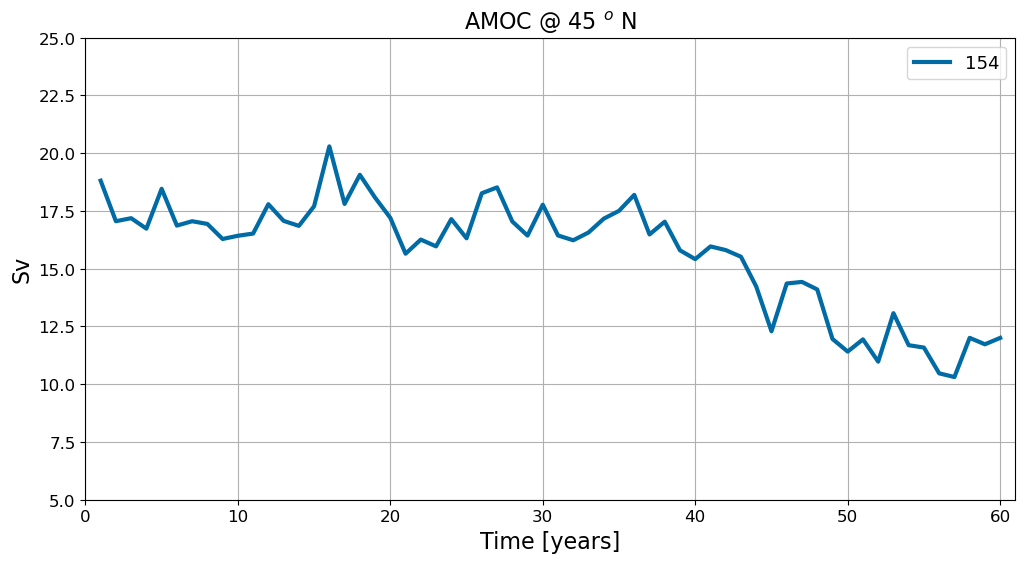
# plot
fig = plt.figure(figsize=(12, 6))
for i in range(pnum):
plt.plot(np.arange(len(ds[i].time))+1 ,ds[i]['amoc_45'].values,
label=label[i], lw=3)
plt.title('AMOC @ 45 $^o$ N', fontsize=16)
plt.ylim(5,25)
plt.xlim(0,1+len(ds[0].time))
plt.xlabel('Time [years]', fontsize=16); plt.ylabel('Sv', fontsize=16)
plt.legend(fontsize=13, ncol=2);
plt.grid()
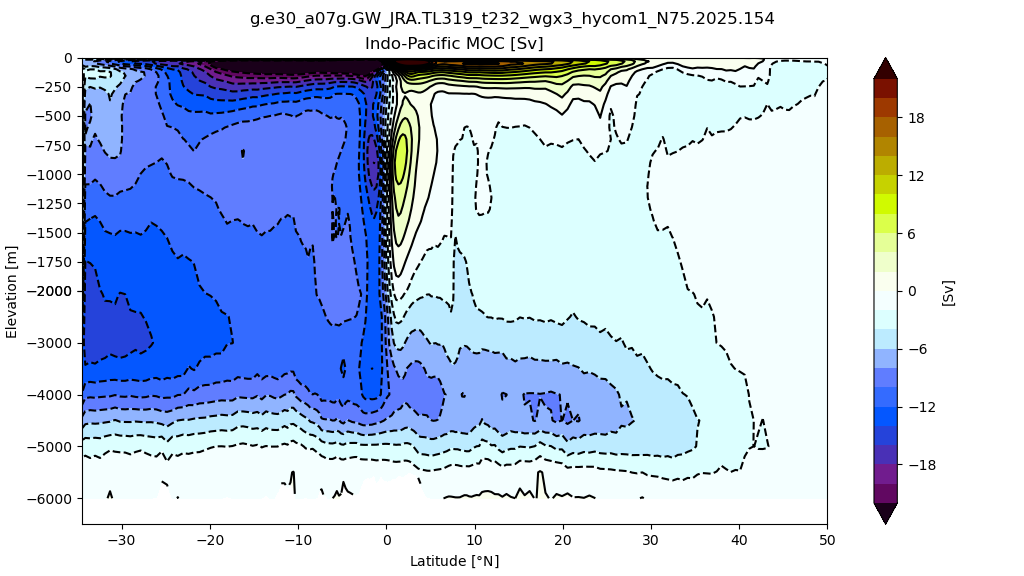
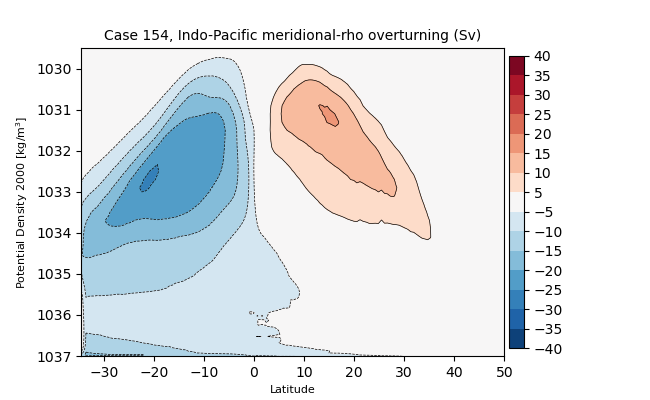
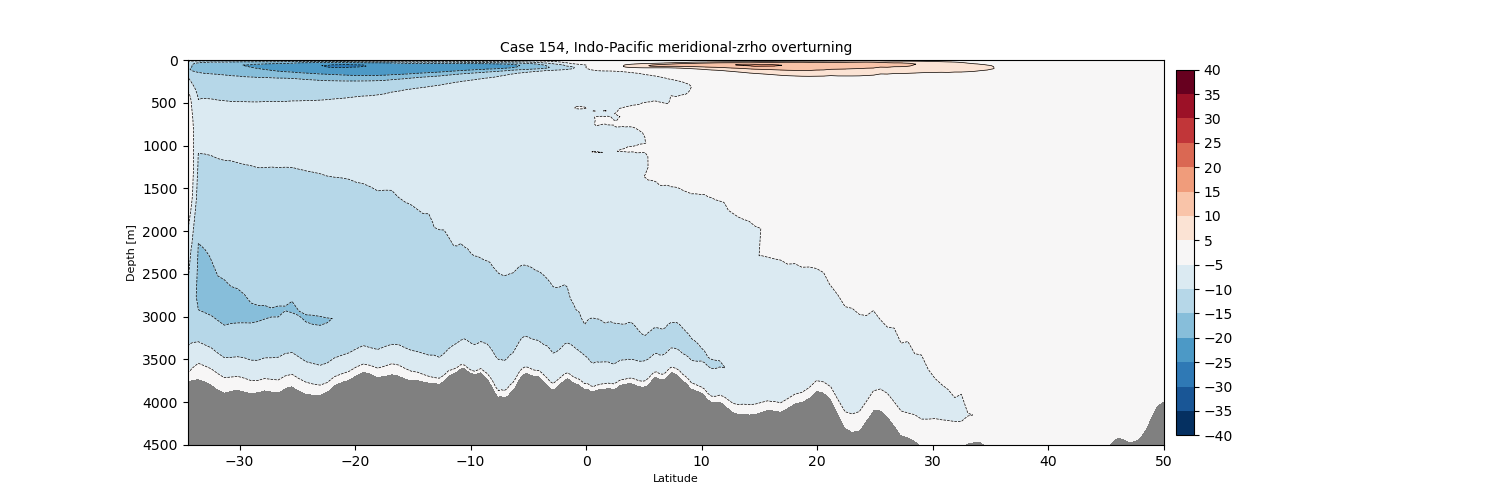
Indo-Pacific#
z#
for i in range(len(casename)):
png_files = glob.glob(ocn_path[i]+'../PNG/MOC/'+casename[i]+'_MOC_IndoPacific.png')
for png_file in png_files:
display(Image(filename=png_file))
sigma2#
for i in range(len(casename)):
png_files = glob.glob(ocn_path[i]+'../PNG/MOC/'+casename[i]+'_MOC_sigma2_IndoPacific.png')
for png_file in png_files:
display(Image(filename=png_file))
zrho#
for i in range(len(casename)):
png_files = glob.glob(ocn_path[i]+'../PNG/MOC/'+casename[i]+'_MOC_zrho_IndoPacific.png')
for png_file in png_files:
display(Image(filename=png_file))
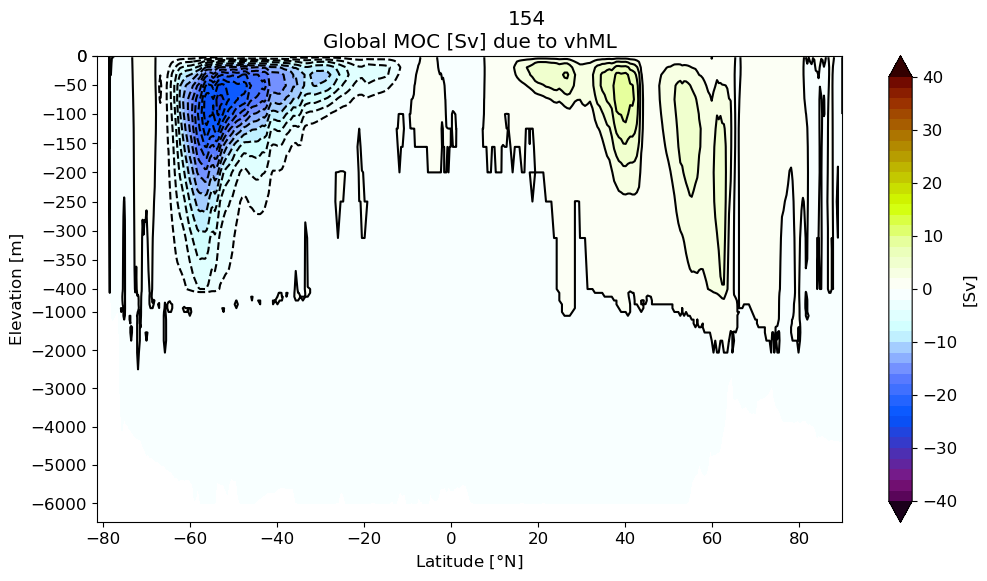
Submesoscale-induced Global MOC#
z#
for i in range(pnum):
# create a ndarray subclass
class C(np.ndarray): pass
varName = 'moc_FFH'
psiPlot = np.ma.masked_invalid(ds[i][varName].values)
tmp = psiPlot[:].filled(0.)
VHmod = tmp.view(C)
VHmod.units = 'Sv'
# Global MOC
m6plot.setFigureSize([16,9],576,debug=False)
axis = plt.gca()
cmap = plt.get_cmap('dunnePM')
z = Zmod[i].min(axis=-1)
#yy = y[1:,:].max(axis=-1)+0*z
yy = grd[i].geolat_c[:,:].max(axis=-1)+0*z
ci=m6plot.pmCI(0.,40.,2.)
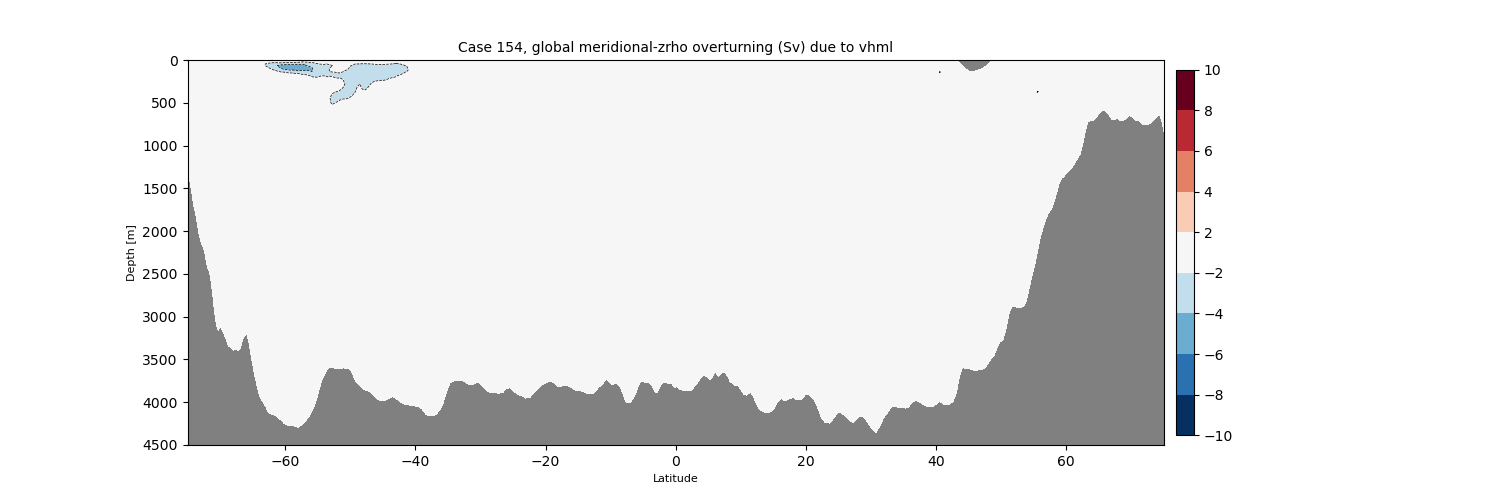
plotPsi(yy, z, psiPlot, ci, 'Global MOC [Sv] due to vhML', zval=[0.,-400.,-6500.])
plt.xlabel(r'Latitude [$\degree$N]')
plt.suptitle(label[i])
plt.gca().invert_yaxis()
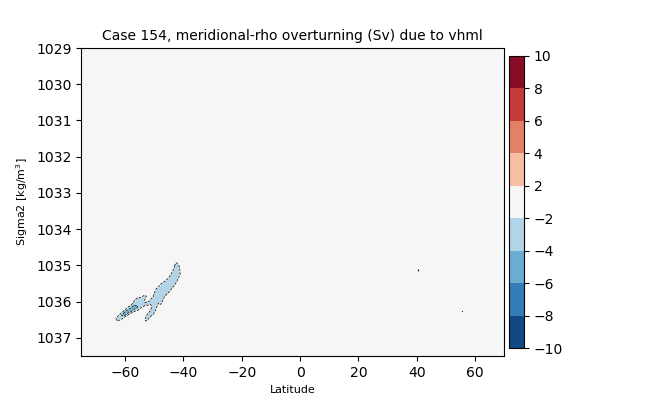
sigma2#
for i in range(len(casename)):
png_files = glob.glob(ocn_path[i]+'../PNG/MOC/'+casename[i]+'_MOC_sigma2_global_vhml.png')
for png_file in png_files:
display(Image(filename=png_file))
zrho#
for i in range(len(casename)):
png_files = glob.glob(ocn_path[i]+'../PNG/MOC/'+casename[i]+'_MOC_zrho_global_vhml.png')
for png_file in png_files:
display(Image(filename=png_file))
Eddy(GM)-induced Global MOC#
z#
for i in range(pnum):
# create a ndarray subclass
class C(np.ndarray): pass
varName = 'moc_GM'
psiPlot = np.ma.masked_invalid(ds[i][varName].values)
tmp = psiPlot[:].filled(0.)
VHmod = tmp.view(C)
VHmod.units = 'Sv'
# Global MOC
m6plot.setFigureSize([16,9],576,debug=False)
axis = plt.gca()
cmap = plt.get_cmap('dunnePM')
z = Zmod[i].min(axis=-1)
yy = grd[0].geolat_c[:,:].max(axis=-1)+0*z
ci=m6plot.pmCI(0.,20.,1.)
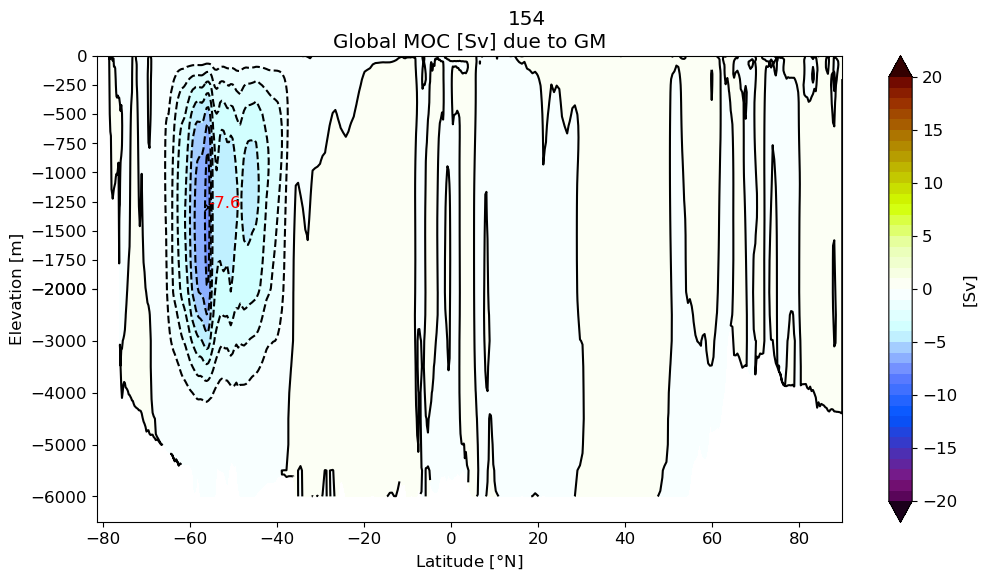
plotPsi(yy, z, psiPlot, ci, 'Global MOC [Sv] due to GM')
plt.xlabel(r'Latitude [$\degree$N]')
plt.suptitle(label[i])
findExtrema(yy, z, psiPlot, min_lat=-65., max_lat=-30, mult=-1.)
plt.gca().invert_yaxis()
sigma2#
for i in range(len(casename)):
png_files = glob.glob(ocn_path[i]+'../PNG/MOC/'+casename[i]+'_MOC_sigma2_global_vhGM.png')
for png_file in png_files:
display(Image(filename=png_file))
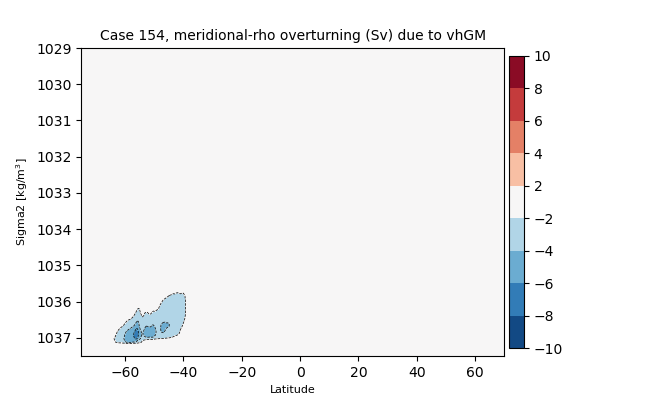
zrho#
for i in range(len(casename)):
png_files = glob.glob(ocn_path[i]+'../PNG/MOC/'+casename[i]+'_MOC_zrho_global_vhGM.png')
for png_file in png_files:
display(Image(filename=png_file))