T & S biases#
%load_ext autoreload
%autoreload 2
%%capture
# comment above line to see details about the run(s) displayed
from misc import *
import nc_time_axis
%matplotlib inline
Drifts#
# these are some of the regions for the drift plots
reg_mom = ['Global', 'PersianGulf', 'RedSea', 'BlackSea', 'MedSea', 'BalticSea',
'HudsonBay', 'Arctic', 'PacificOcean', 'AtlanticOcean', 'IndianOcean',
'SouthernOcean', 'LabSea', 'BaffinBay', 'EGreenlandIceland',
'GulfOfMexico', 'Maritime', 'SouthernOcean60S']
Potential temperature#
nr=1
nc=1
fs=(9,5.5)
matplotlib.rcParams.update({'font.size': 17})
def make_temp_drift_plot(m='Global', vmax=1.5, splitscale = [0., -500., -4000]):
vmin=-vmax
for path, case, i in zip(ocn_path, casename, range(len(casename))):
fig, ax = plt.subplots(nrows=nr, ncols=nc, figsize=fs, sharex=False, sharey=True)
plt.suptitle(str(m)+', Potential Temperature bias [C]', fontsize=14)
ds_mom = xr.open_dataset(path+case+'_thetao_drift.nc').sel(time=slice('0001-01-01', end_date))
dummy_mom = np.ma.masked_invalid(ds_mom.sel(region=m).thetao_drift.values)
ztplot(dummy_mom, ds_mom.time.values, ds_mom.z_l.values*-1, ignore=np.nan, splitscale=splitscale,
contour=True, axis=ax , title=label[i], extend='both', colormap='dunnePM',
autocenter=True, tunits='Year', show=False, clim=(vmin, vmax))
plt.subplots_adjust(top = 0.8)
plt.tight_layout()
return
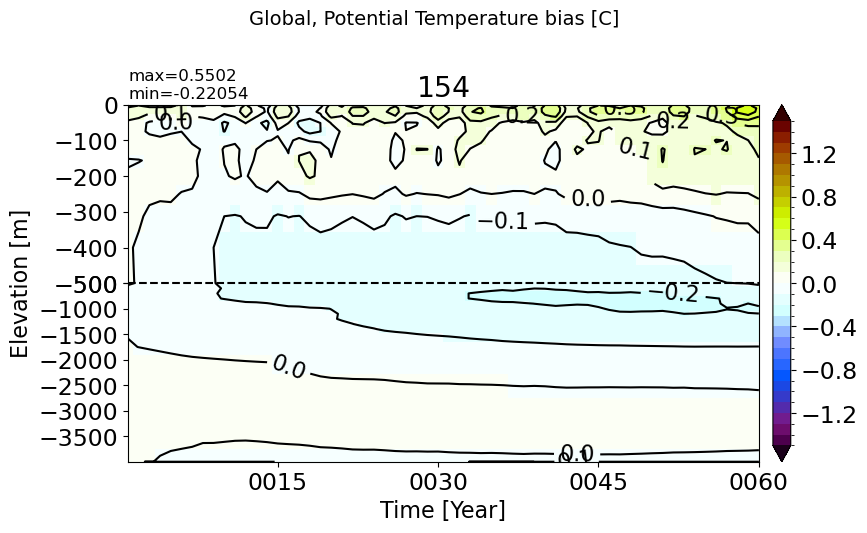
Global#
make_temp_drift_plot()
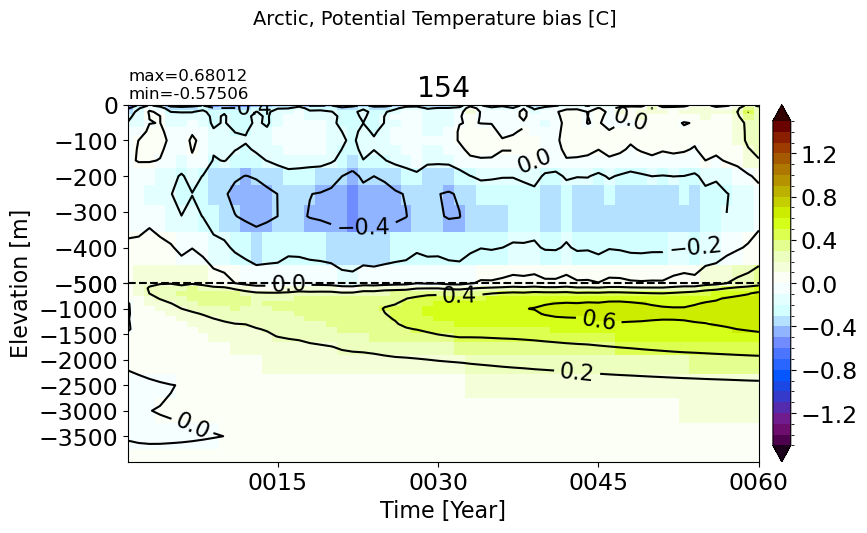
Arctic#
make_temp_drift_plot(m='Arctic')
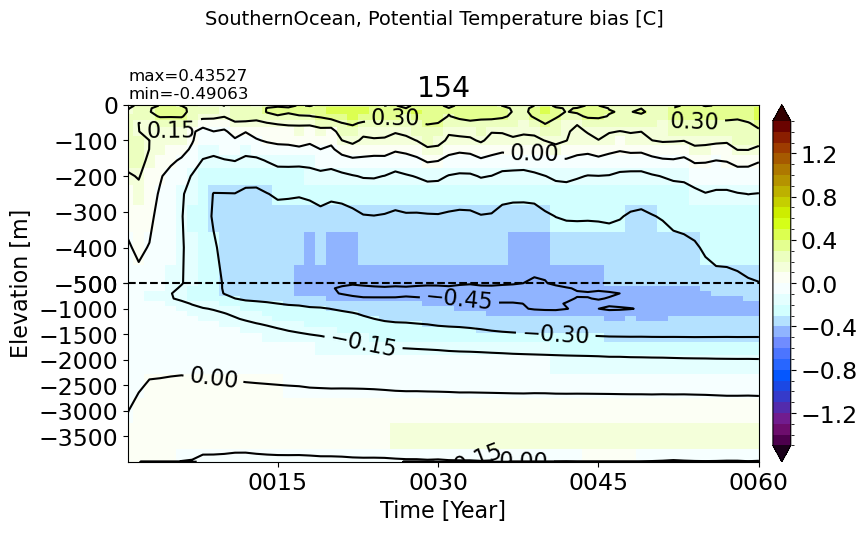
SouthernOcean#
make_temp_drift_plot(m='SouthernOcean')
SouthernOcean60S#
make_temp_drift_plot(m='SouthernOcean60S')
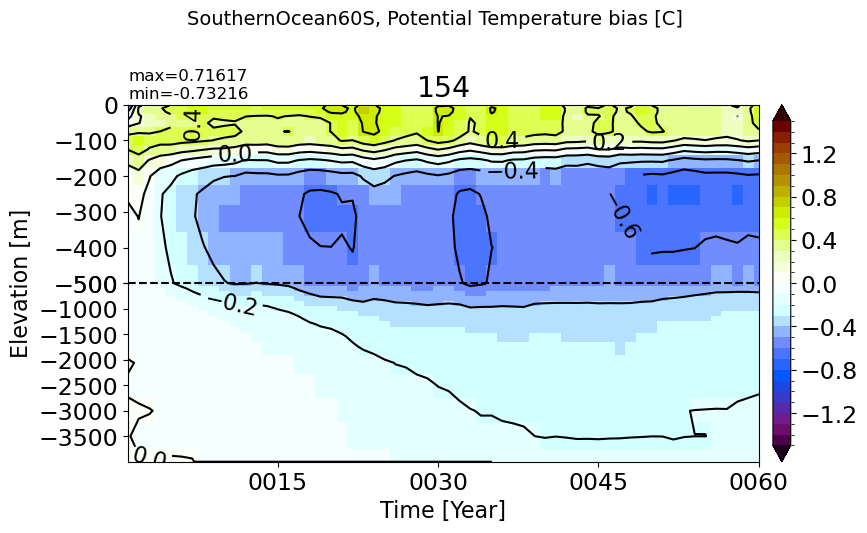
PacificOcean#
make_temp_drift_plot(m='PacificOcean')
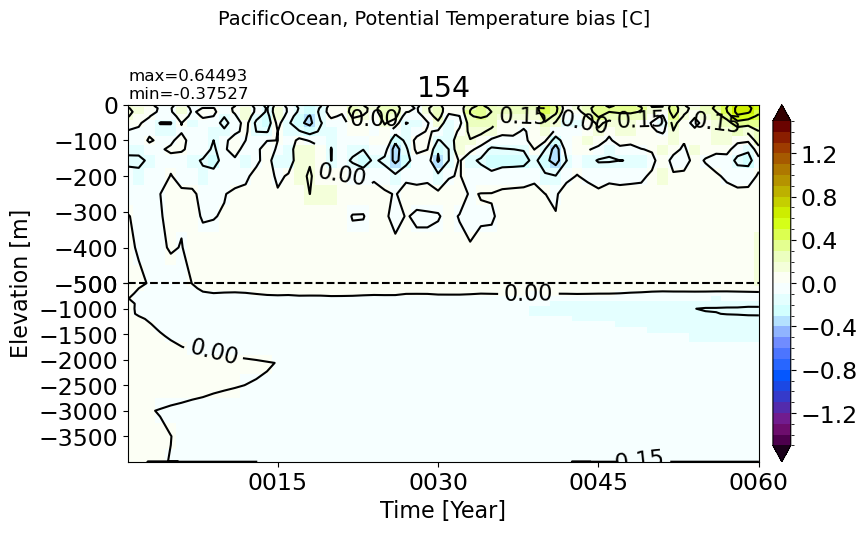
AtlanticOcean#
make_temp_drift_plot(m='AtlanticOcean')
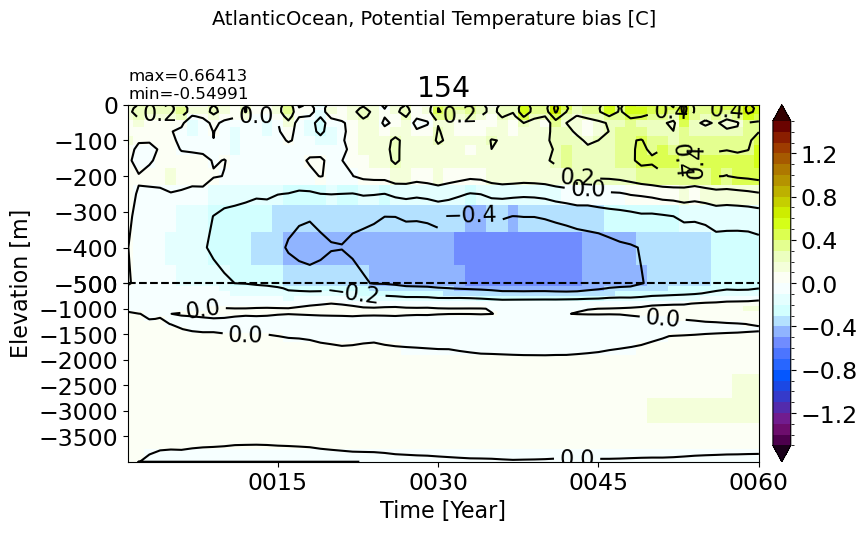
IndianOcean#
make_temp_drift_plot(m='IndianOcean')
Maritime#
make_temp_drift_plot(m='Maritime')
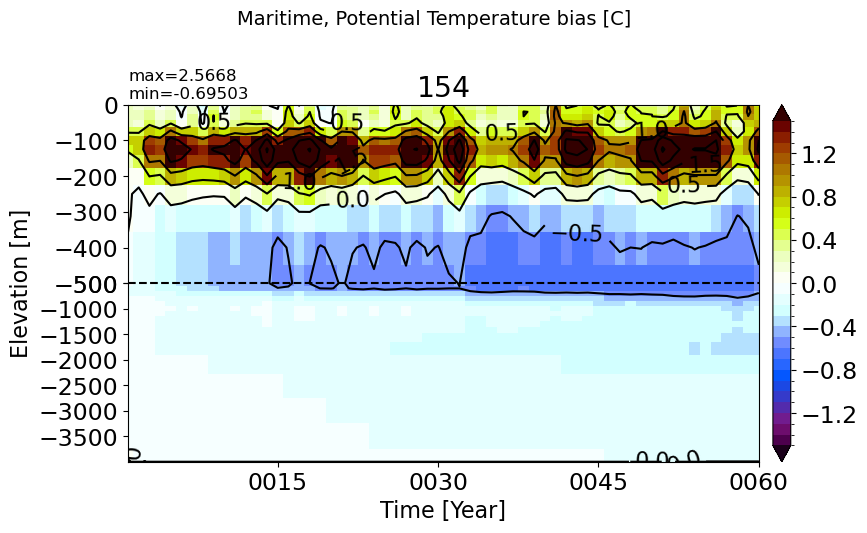
BalticSea#
make_temp_drift_plot(m='BalticSea', splitscale = [0., -100., -400])
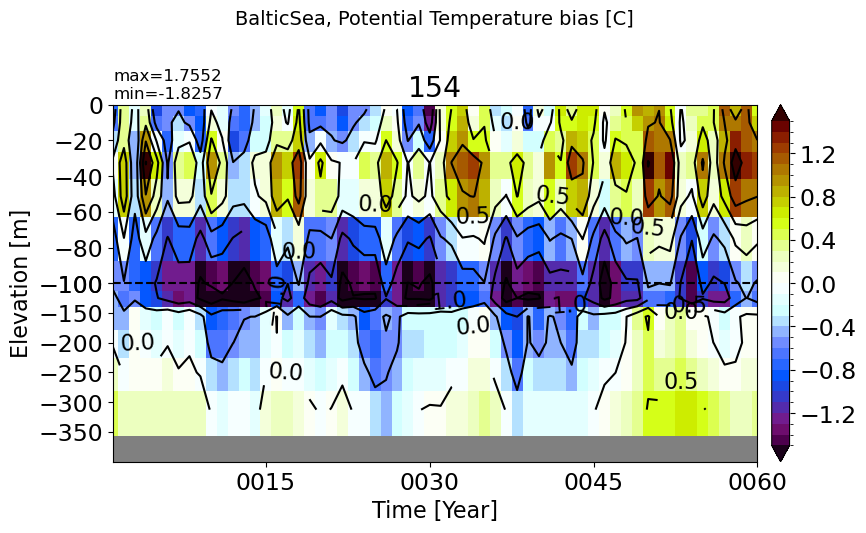
PersianGulf#
make_temp_drift_plot(m='PersianGulf', splitscale = [0., -100., -70])
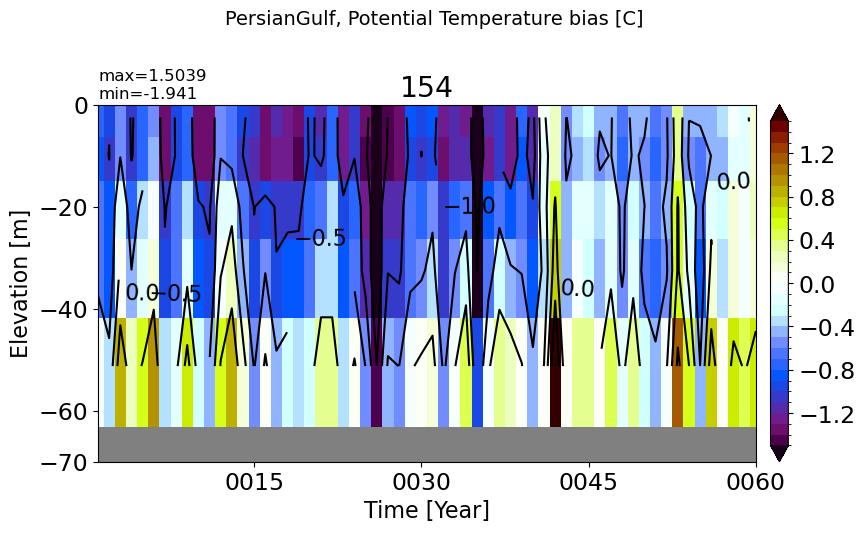
RedSea#
make_temp_drift_plot(m='RedSea', splitscale = [0., -200., -1200])
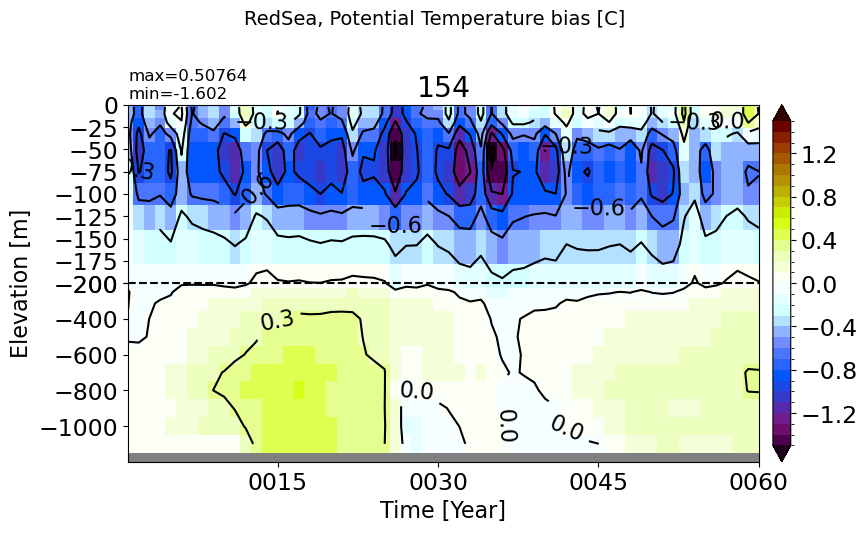
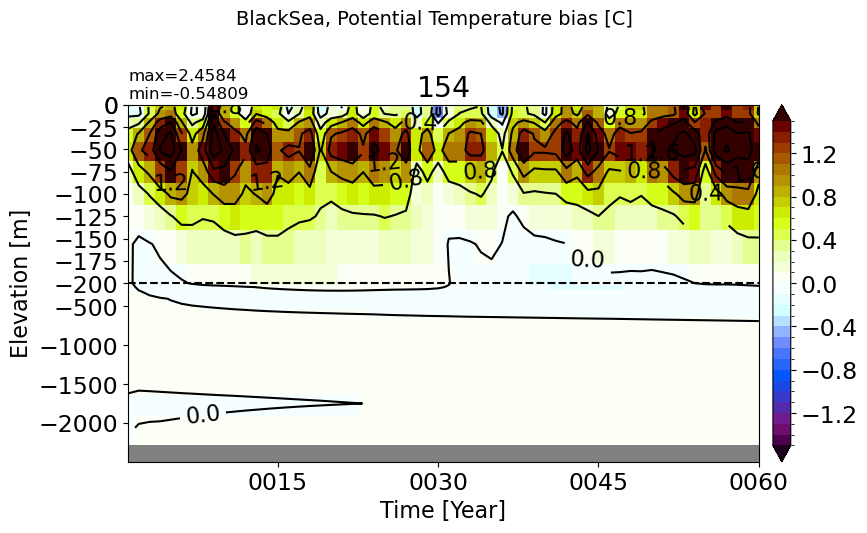
BlackSea#
make_temp_drift_plot(m='BlackSea', splitscale = [0., -200., -2500])
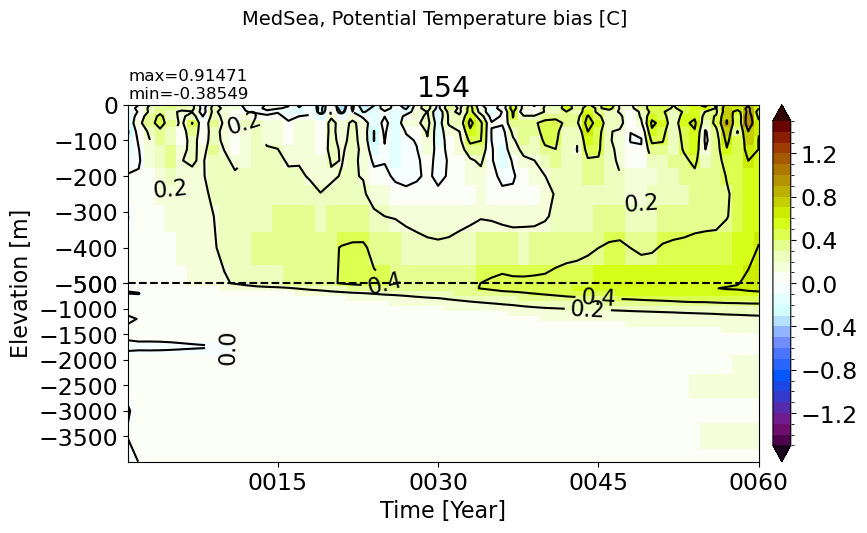
MedSea#
make_temp_drift_plot(m='MedSea')
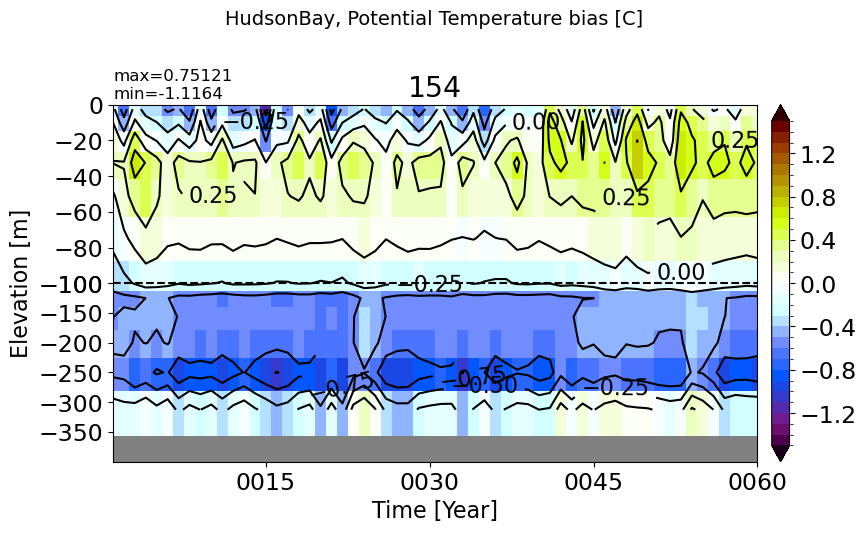
HudsonBay#
make_temp_drift_plot(m='HudsonBay', splitscale = [0., -100., -400])
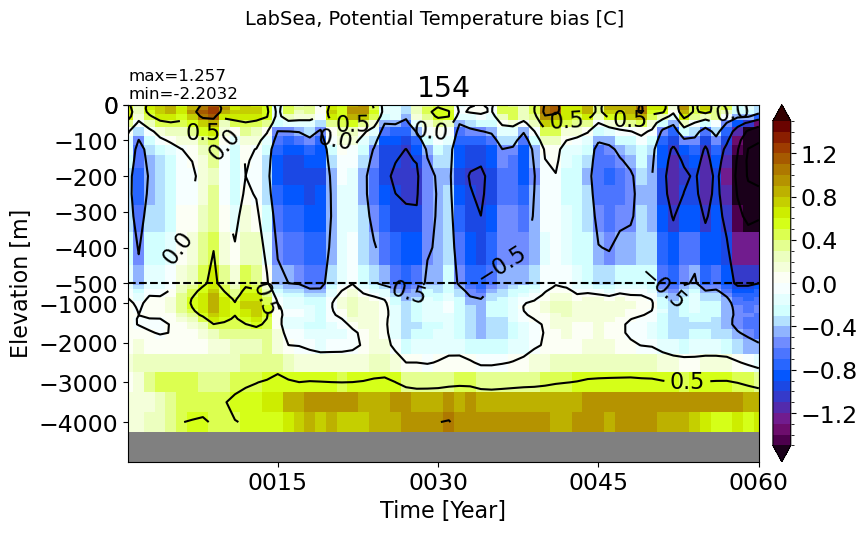
LabSea#
make_temp_drift_plot(m='LabSea', splitscale = [0., -500., -5000])
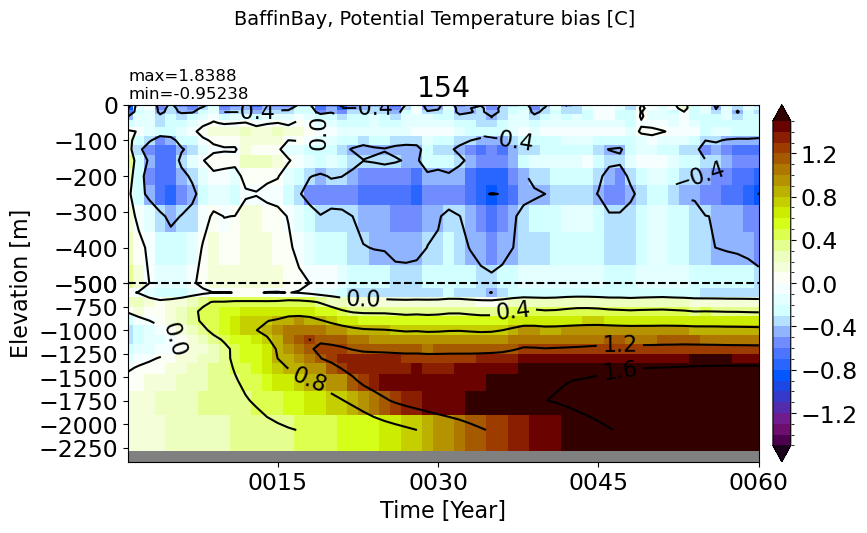
BaffinBay#
make_temp_drift_plot(m='BaffinBay', splitscale = [0., -500., -2400])
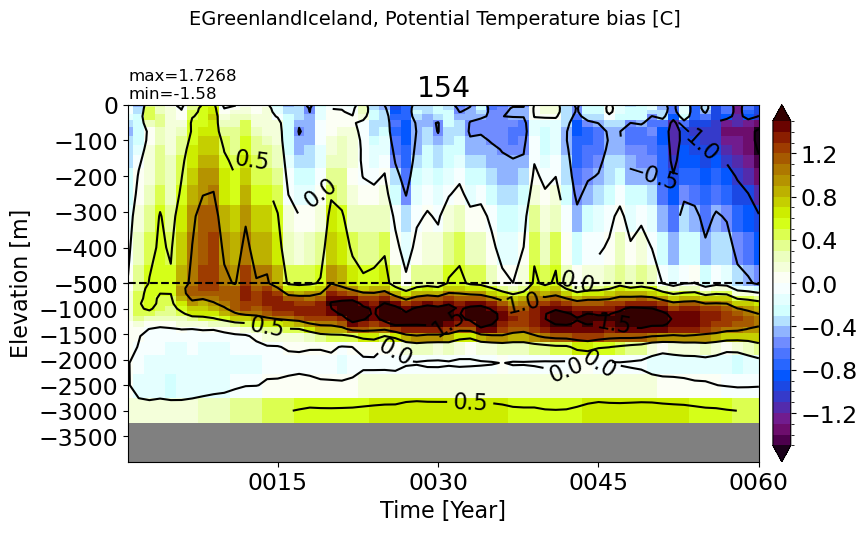
EGreenlandIceland#
make_temp_drift_plot(m='EGreenlandIceland')
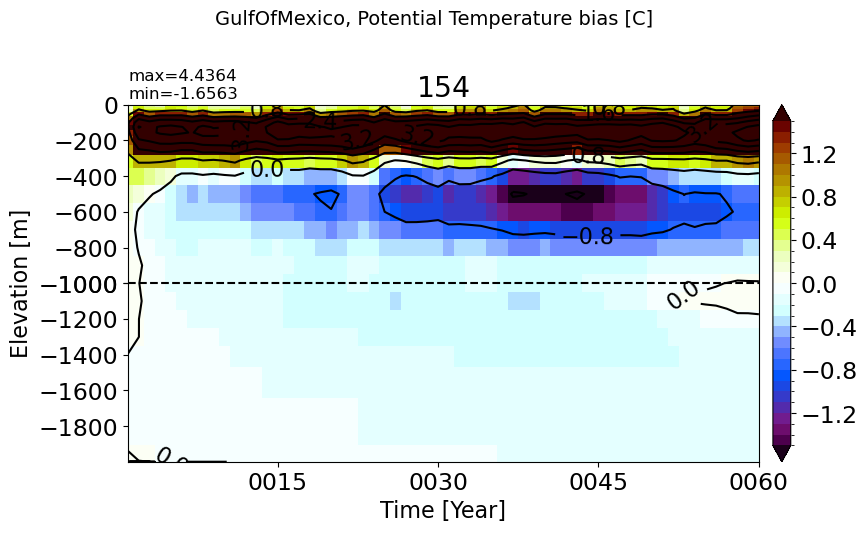
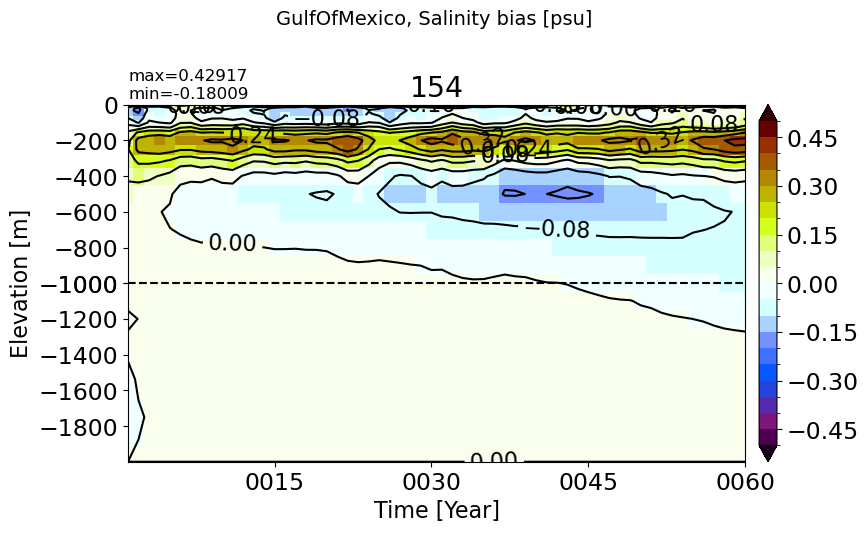
GulfOfMexico#
make_temp_drift_plot(m='GulfOfMexico', splitscale = [0., -1000., -2000])
Salinity#
def make_salt_drift_plot(m='Global', vmax=0.1, splitscale=[0., -500., -4000]):
vmin=-vmax
for path, case, i in zip(ocn_path, casename, range(len(casename))):
fig, ax = plt.subplots(nrows=nr, ncols=nc, figsize=fs, sharex=False, sharey=True)
plt.suptitle(str(m)+', Salinity bias [psu]', fontsize=14)
ds_mom = xr.open_dataset(path+case+'_so_drift.nc').sel(time=slice('0001-01-01', end_date))
dummy_mom = np.ma.masked_invalid(ds_mom.sel(region=m).so_drift.values)
ztplot(dummy_mom, ds_mom.time.values, ds_mom.z_l.values*-1, ignore=np.nan, splitscale=splitscale,
contour=True, axis=ax , title=label[i], extend='both', colormap='dunnePM',
autocenter=True, tunits='Year', show=False, clim=(vmin, vmax))
plt.subplots_adjust(top = 0.8)
plt.tight_layout()
return
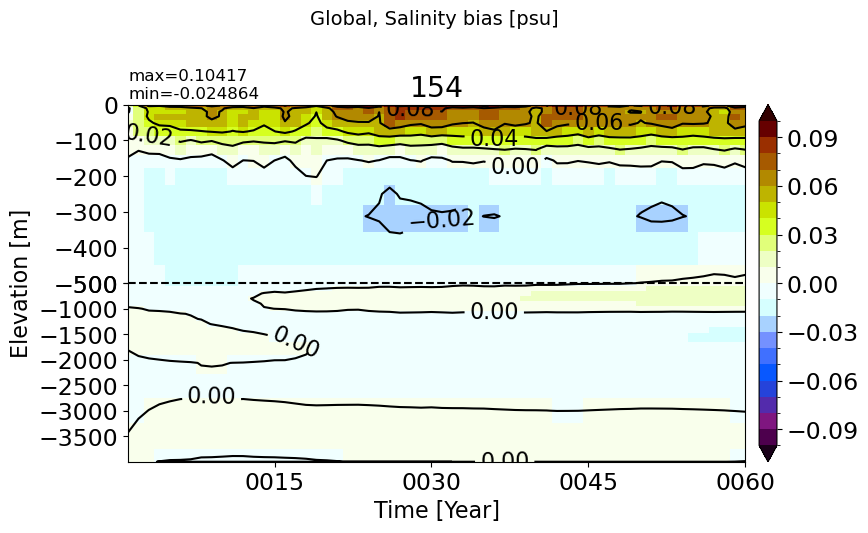
Global#
make_salt_drift_plot()
Arctic#
make_salt_drift_plot(m='Arctic', vmax=0.5)
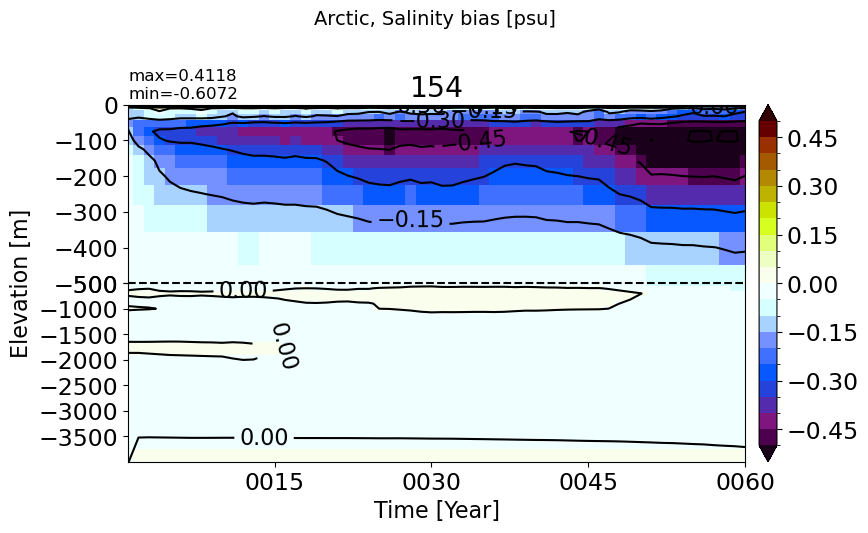
SouthernOcean#
make_salt_drift_plot(m='SouthernOcean')
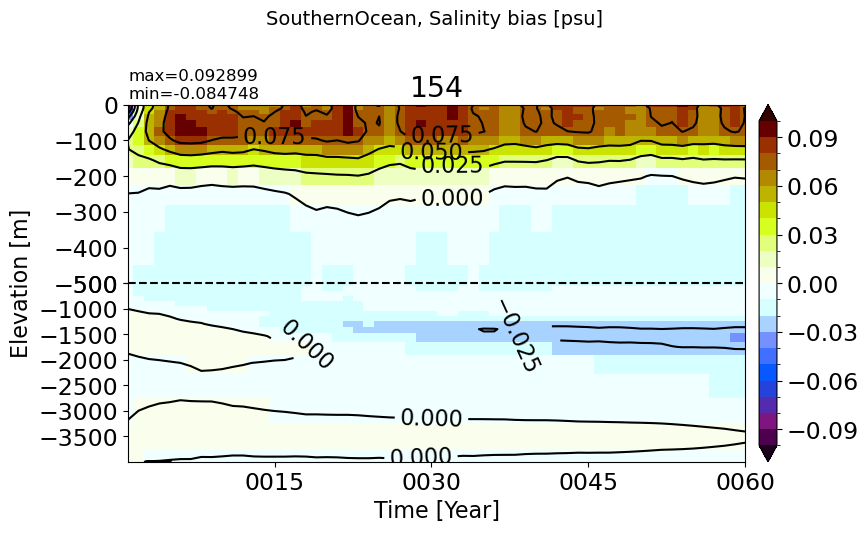
SouthernOcean60S#
make_salt_drift_plot(m='SouthernOcean60S')
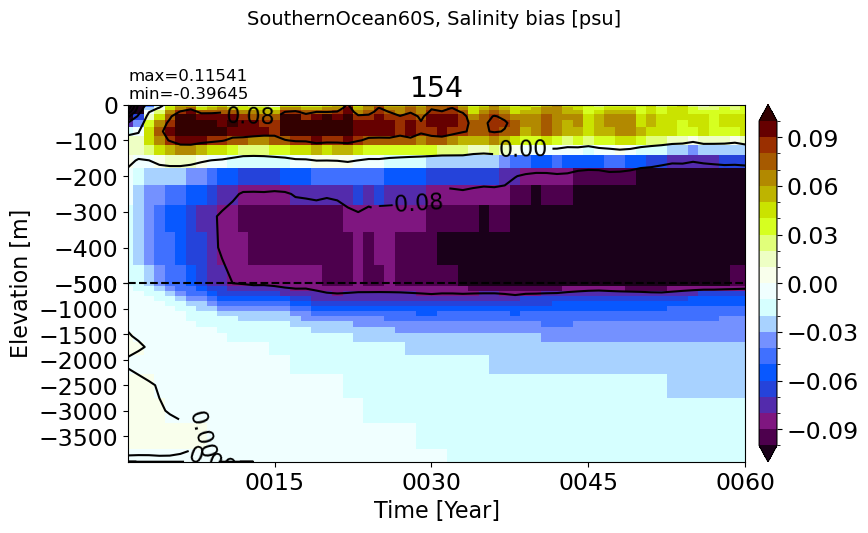
PacificOcean#
make_salt_drift_plot(m='PacificOcean')
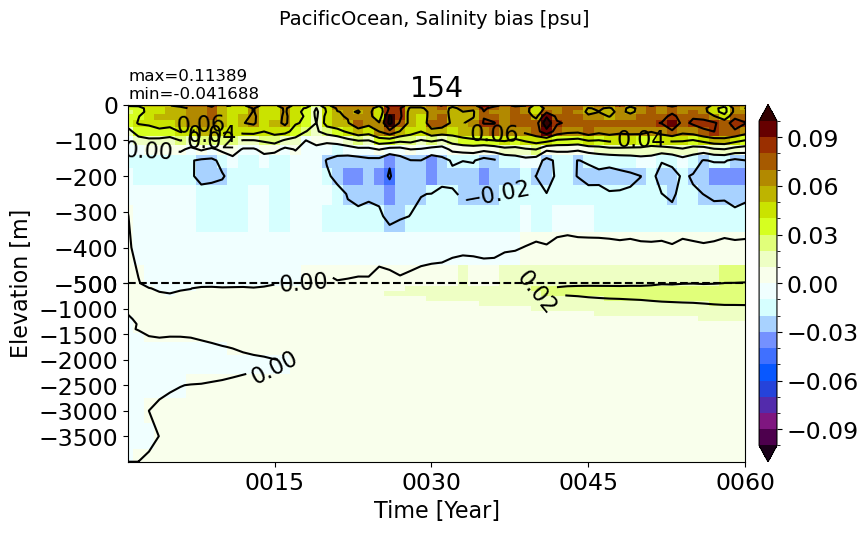
AtlanticOcean#
make_salt_drift_plot(m='AtlanticOcean')
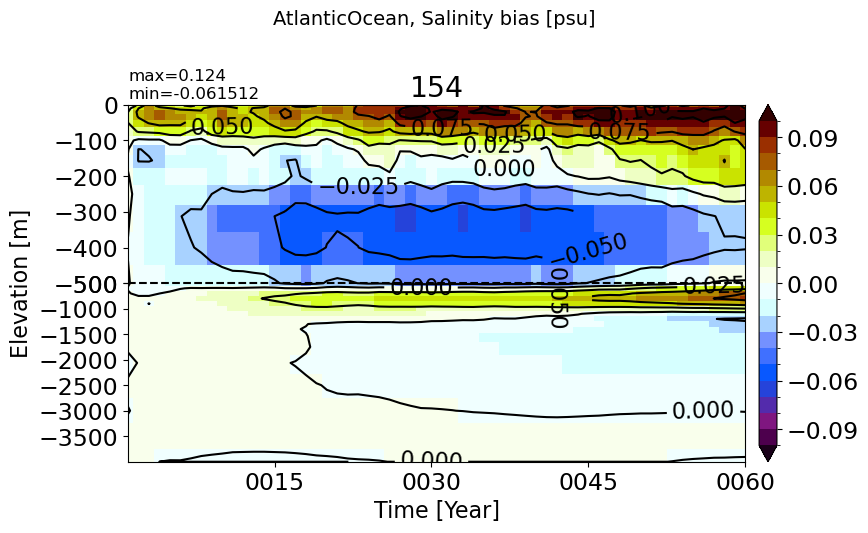
IndianOcean#
make_salt_drift_plot(m='IndianOcean')
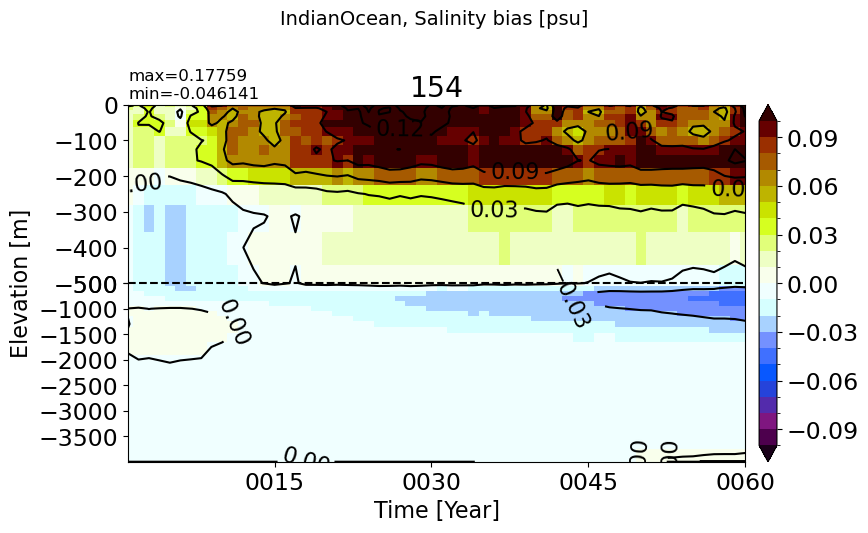
Maritime#
make_salt_drift_plot(m='Maritime')
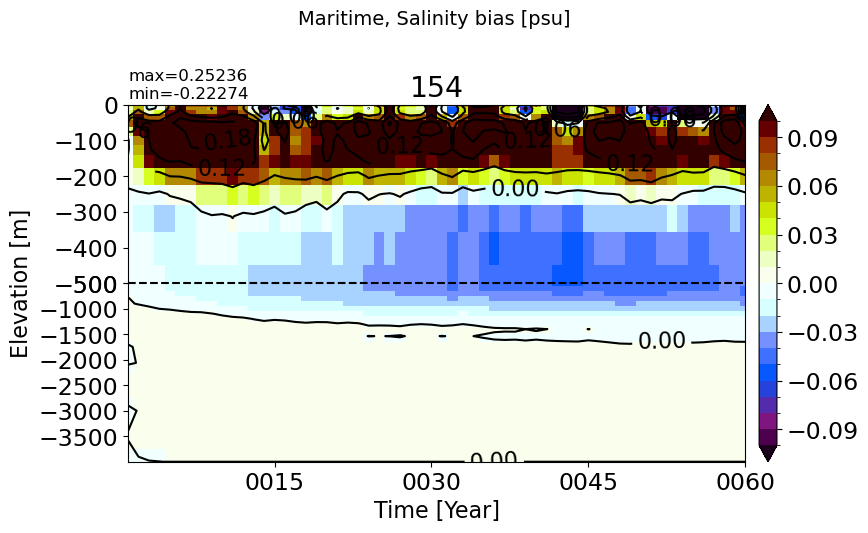
BalticSea#
make_salt_drift_plot(m='BalticSea', vmax=2.5, splitscale = [0., -100., -400])
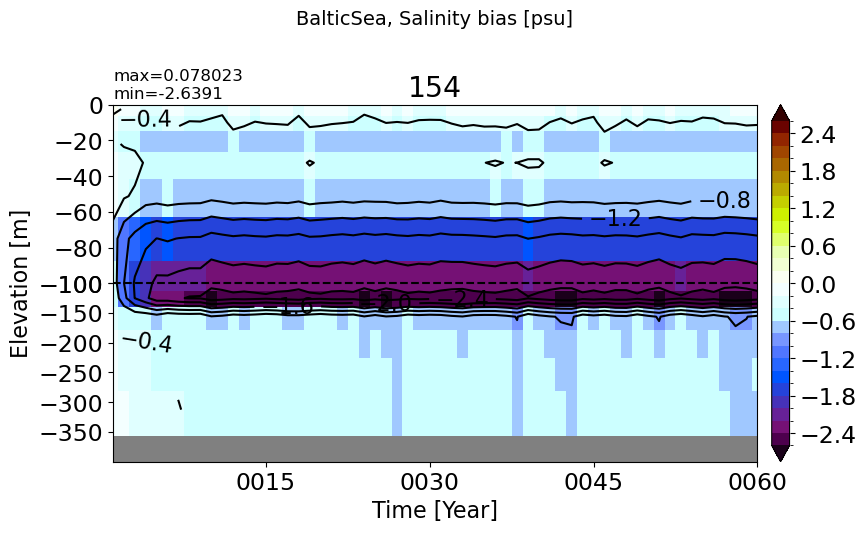
PersianGulf#
make_salt_drift_plot(m='PersianGulf', vmax=0.5, splitscale = [0., -100., -70])
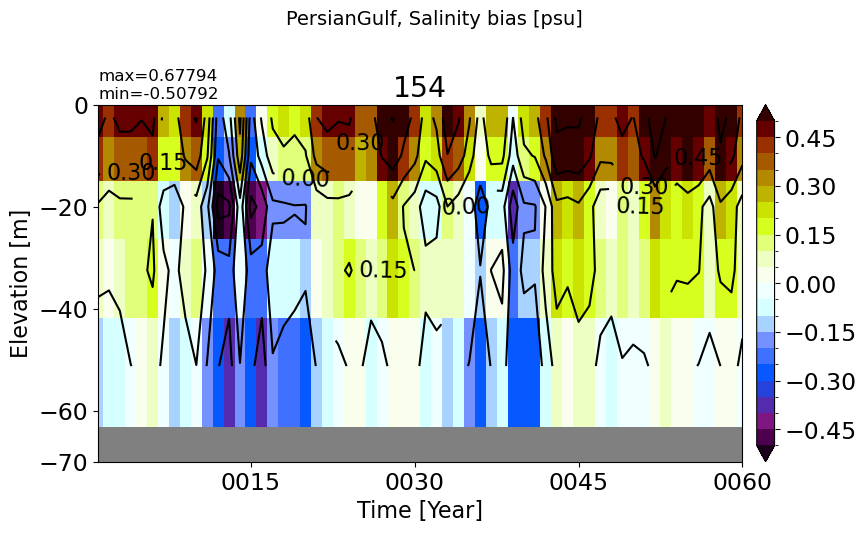
RedSea#
make_salt_drift_plot(m='RedSea', vmax=1.0, splitscale = [0., -200., -1200])
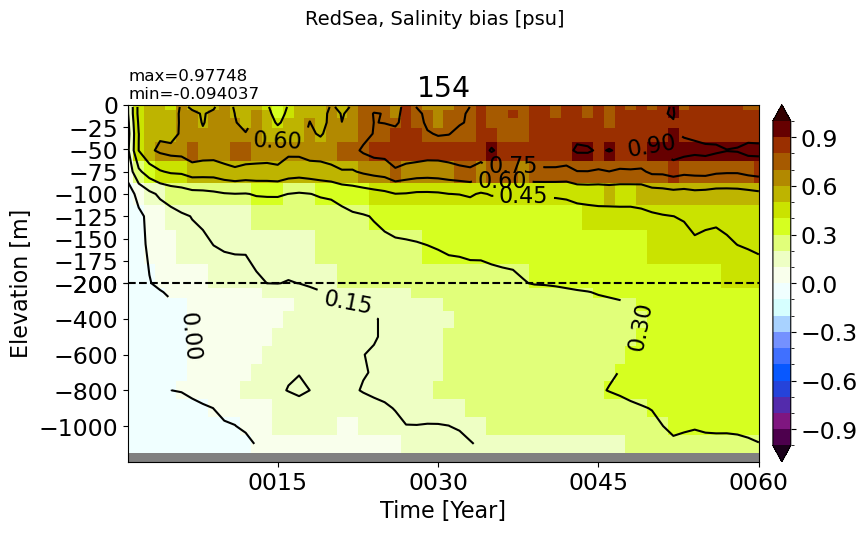
BlackSea#
make_salt_drift_plot(m='BlackSea', vmax=1.5, splitscale = [0., -200., -2500])
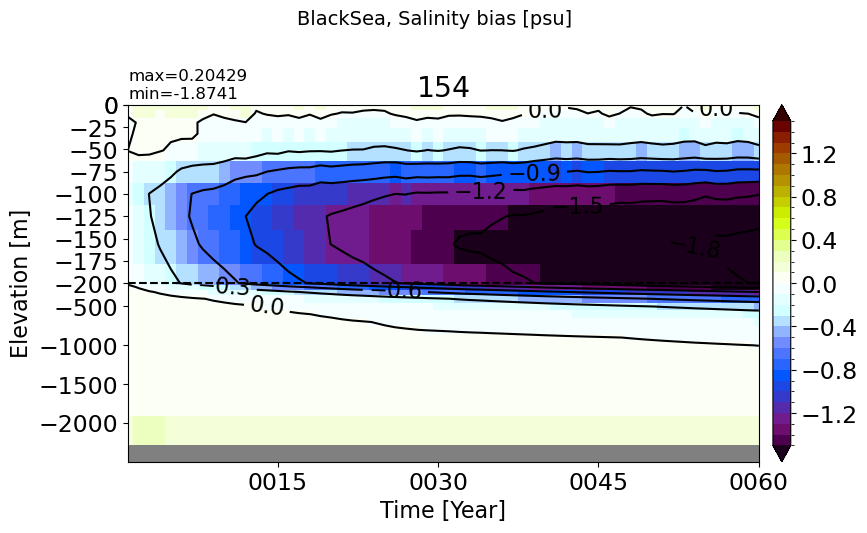
MedSea#
make_salt_drift_plot(m='MedSea')
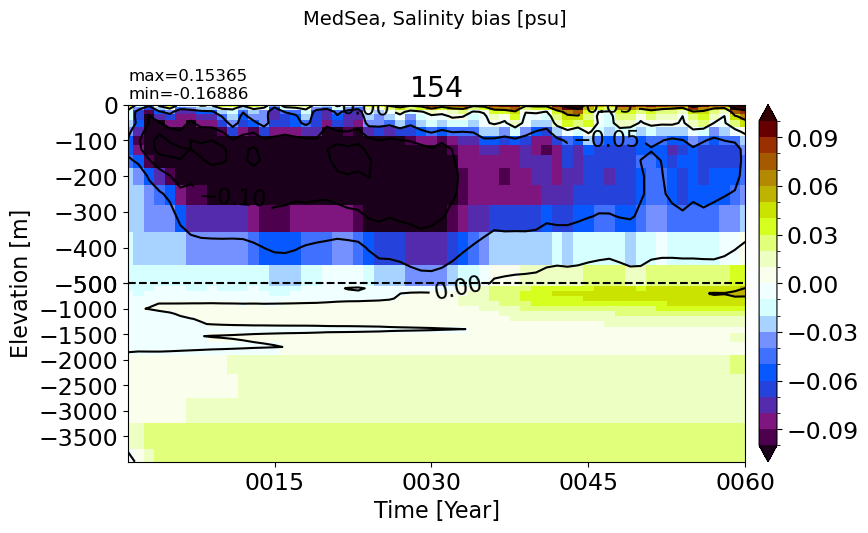
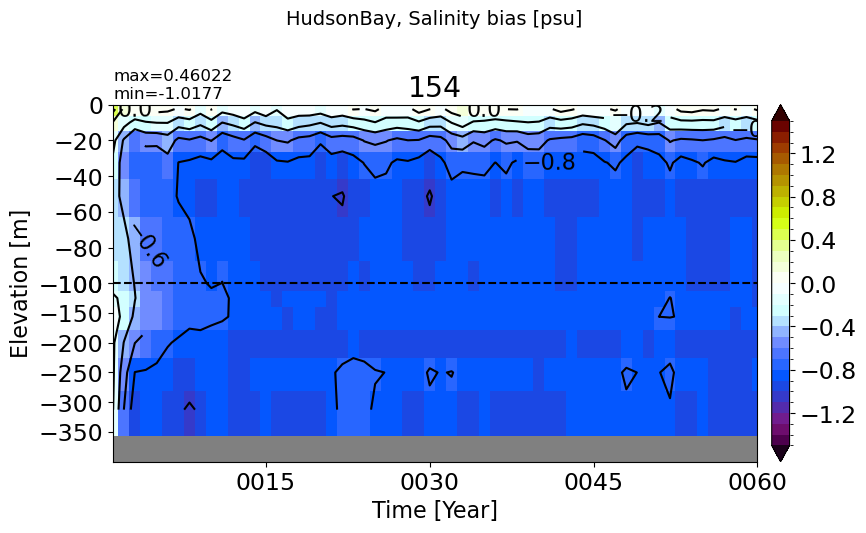
HudsonBay#
make_salt_drift_plot(m='HudsonBay', vmax=1.5, splitscale = [0., -100., -400])
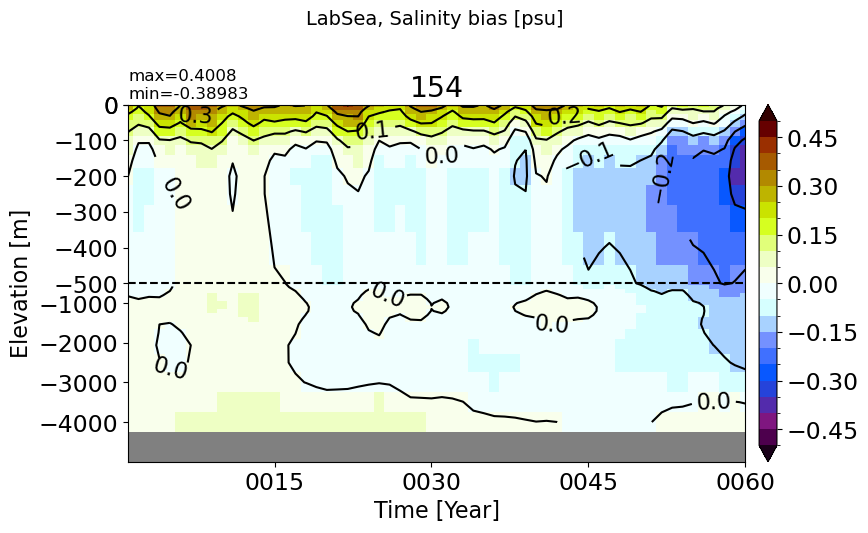
LabSea#
make_salt_drift_plot(m='LabSea', vmax=0.5, splitscale = [0., -500., -5000])
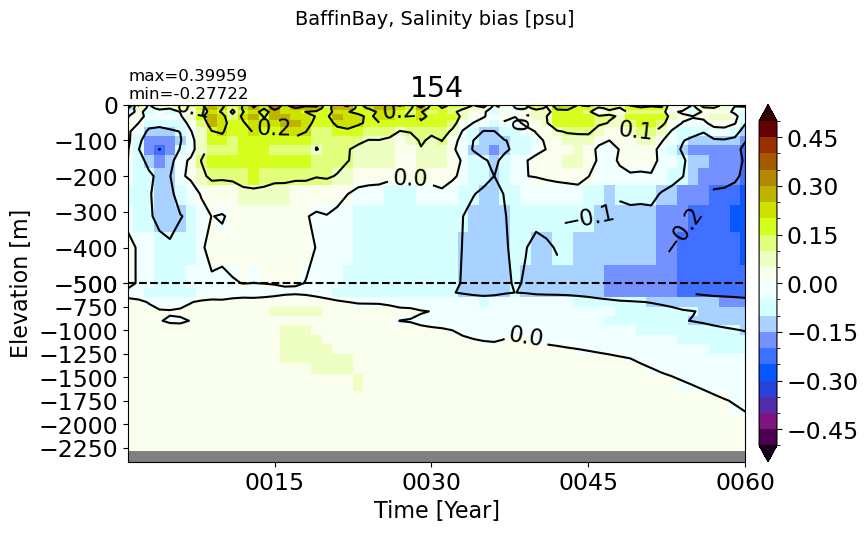
BaffinBay#
make_salt_drift_plot(m='BaffinBay', vmax=0.5, splitscale = [0., -500., -2400])
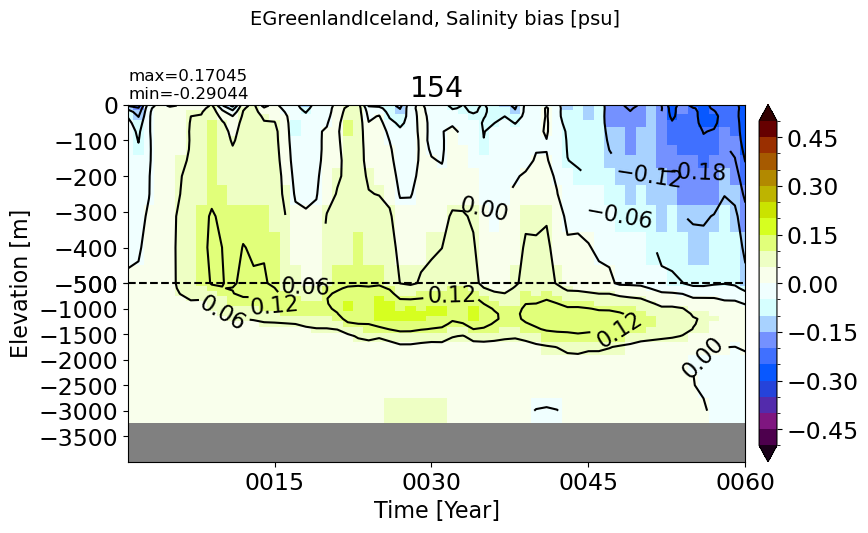
EGreenlandIceland#
make_salt_drift_plot(m='EGreenlandIceland', vmax=0.5)
GulfOfMexico#
make_salt_drift_plot(m='GulfOfMexico', vmax=0.5, splitscale = [0., -1000., -2000])
RMSE#
Potential Temperature#
def make_temp_rmse_plot(m='Global', vmax=3, splitscale = [0., -500., -4000]):
vmin=0
for path, case, i in zip(ocn_path, casename, range(len(casename))):
fig, ax = plt.subplots(nrows=nr, ncols=nc, figsize=fs, sharex=False, sharey=True)
plt.suptitle(str(m)+', Potential Temperature [C] RMS (model - obs)', fontsize=14)
ds_mom = xr.open_dataset(path+case+'_thetao_rmse.nc').sel(time=slice('0001-01-01', end_date))
dummy_mom = np.ma.masked_invalid(ds_mom.sel(region=m).thetao_rms.values)
ztplot(dummy_mom, ds_mom.time.values, ds_mom.z_l.values*-1, ignore=np.nan, splitscale=splitscale,
contour=True, axis=ax , title=label[i], extend='both', colormap='viridis',
autocenter=True, tunits='Year', show=False, clim=(vmin, vmax))
plt.subplots_adjust(top = 0.8)
plt.tight_layout()
return
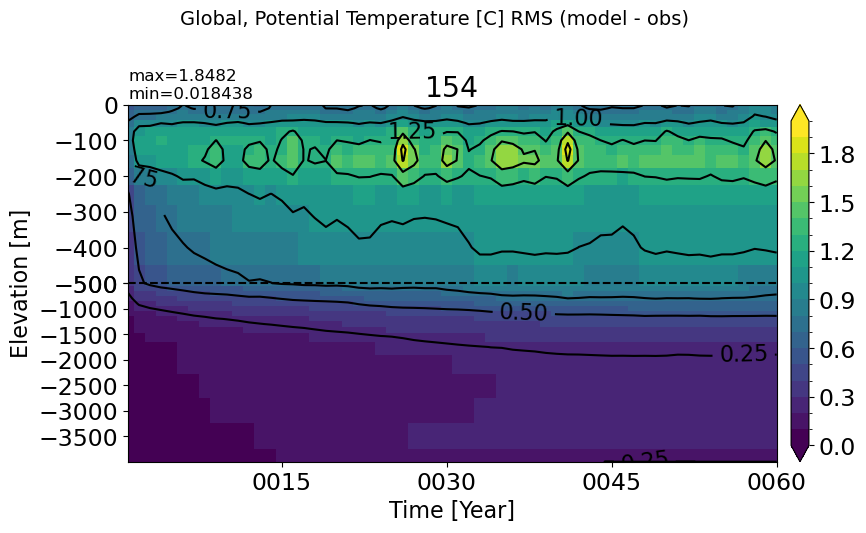
Global#
make_temp_rmse_plot(vmax=2)
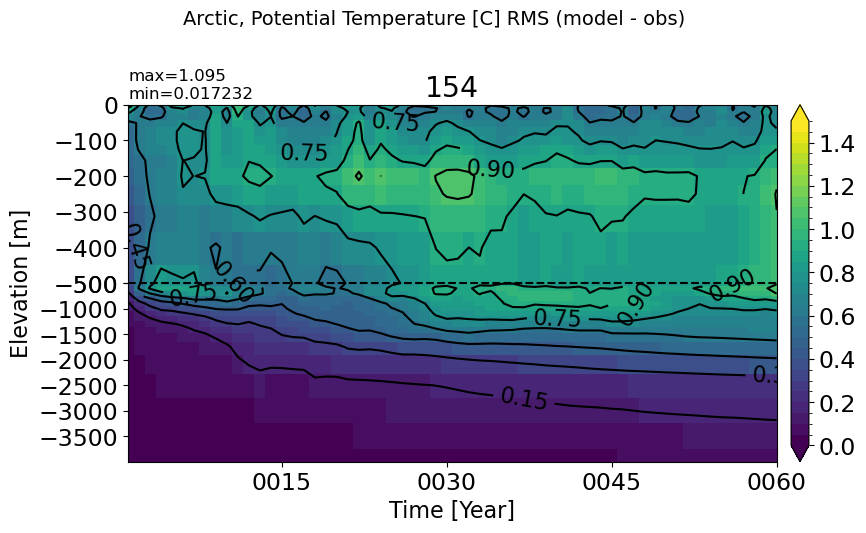
Arctic#
make_temp_rmse_plot(m='Arctic', vmax=1.5)
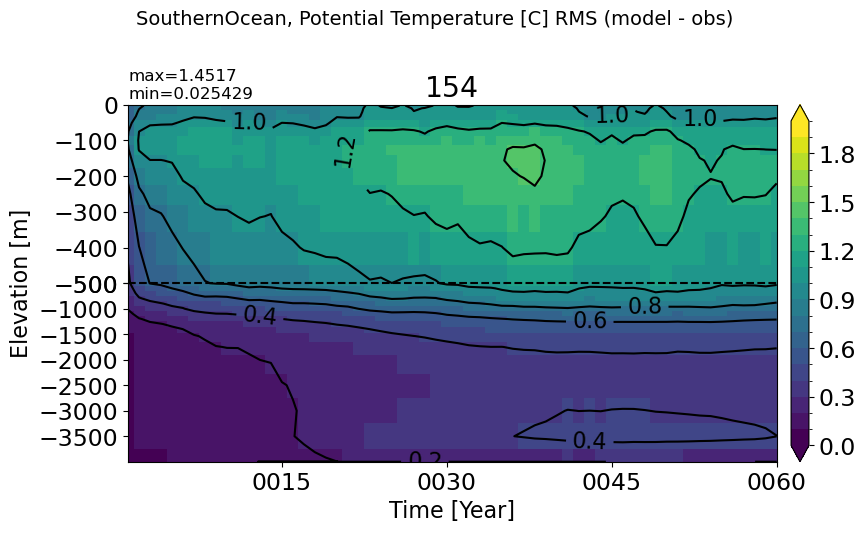
SouthernOcean#
make_temp_rmse_plot(m='SouthernOcean', vmax=2)
SouthernOcean60S#
make_temp_rmse_plot(m='SouthernOcean60S', vmax=2)
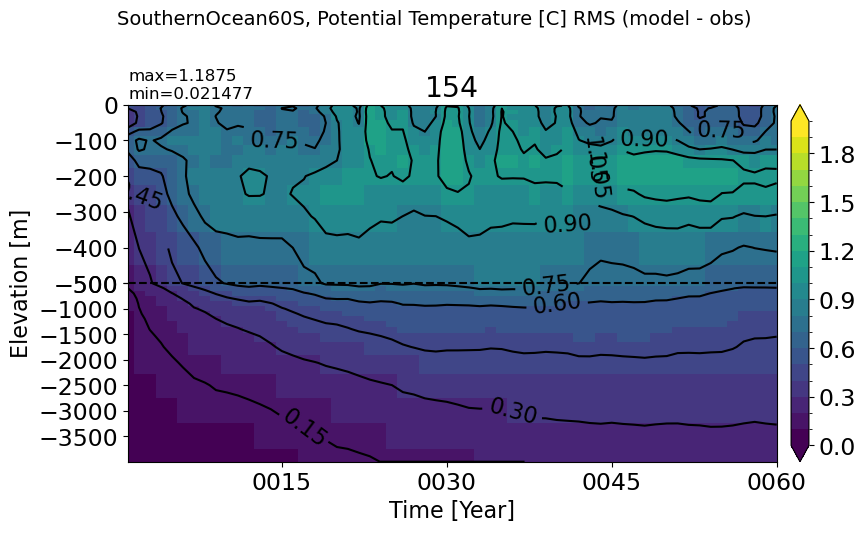
PacificOcean#
make_temp_rmse_plot(m='PacificOcean', vmax=2)
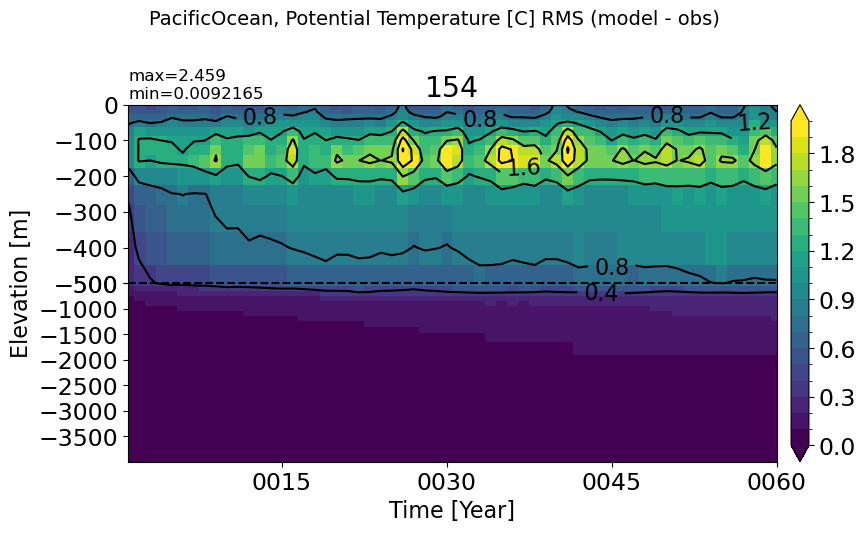
AtlanticOcean#
make_temp_rmse_plot(m='AtlanticOcean', vmax=2)
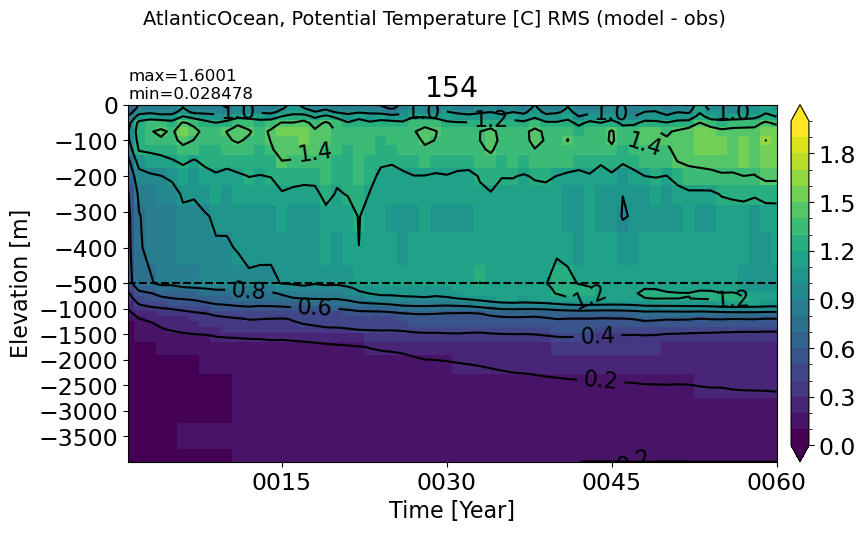
IndianOcean#
make_temp_rmse_plot(m='IndianOcean')
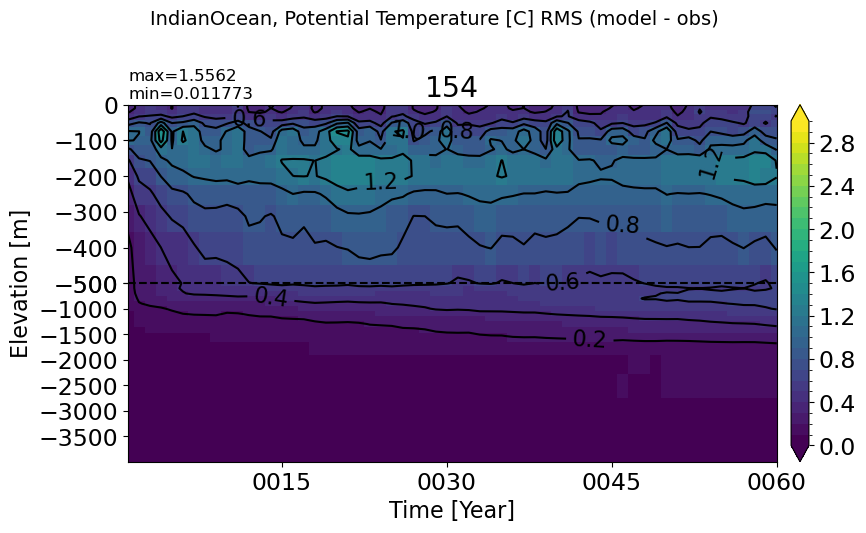
Maritime#
make_temp_rmse_plot(m='Maritime')
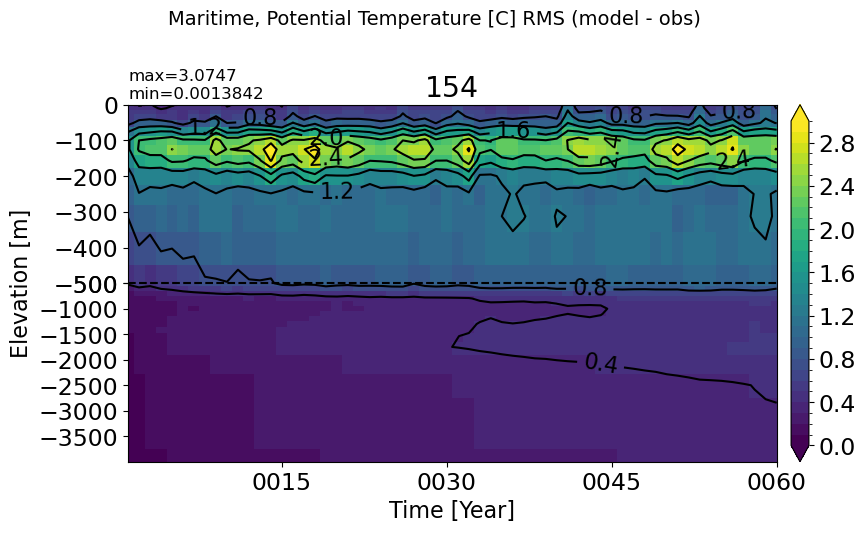
BalticSea#
make_temp_rmse_plot(m='BalticSea', vmax=2, splitscale = [0., -100., -400])
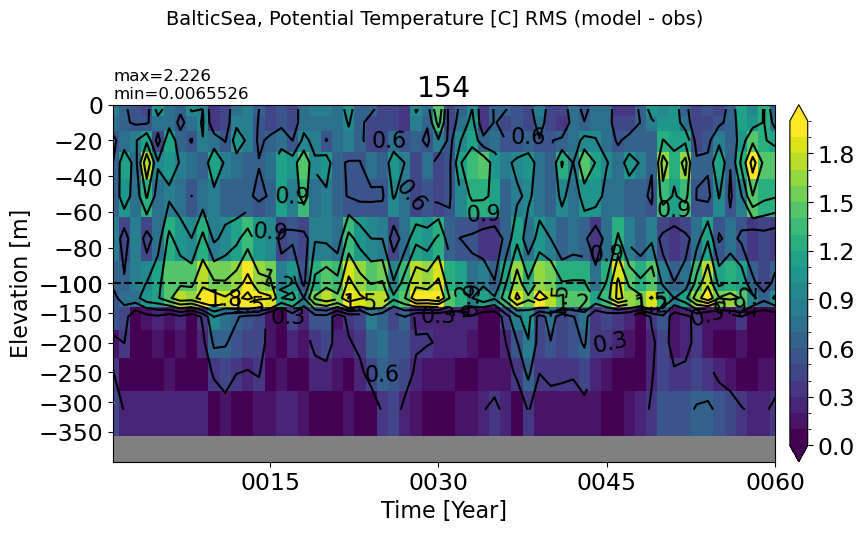
PersianGulf#
make_temp_rmse_plot(m='PersianGulf', vmax=2,splitscale = [0., -100., -70])
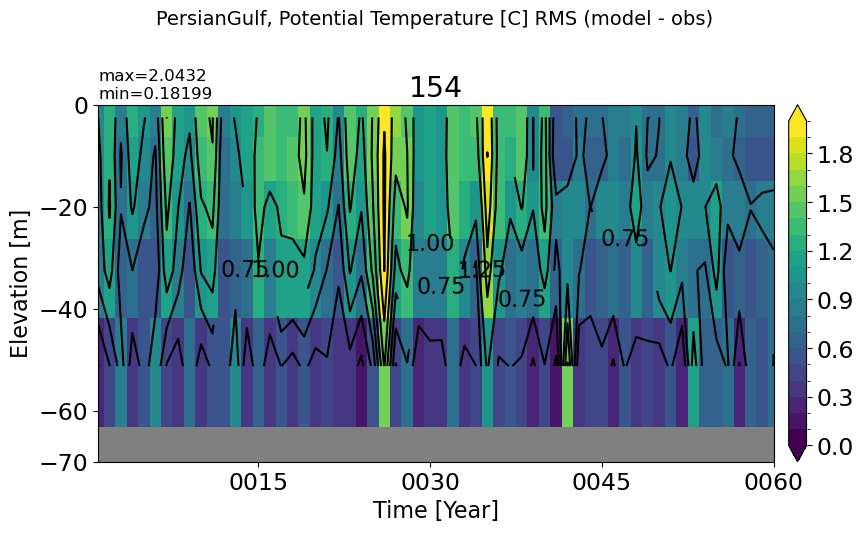
RedSea#
make_temp_rmse_plot(m='RedSea', vmax=2, splitscale = [0., -200., -1200])
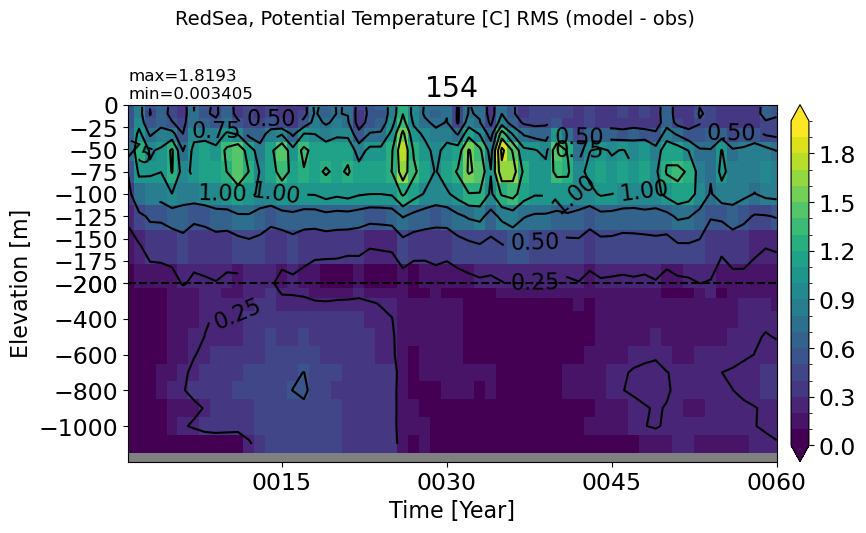
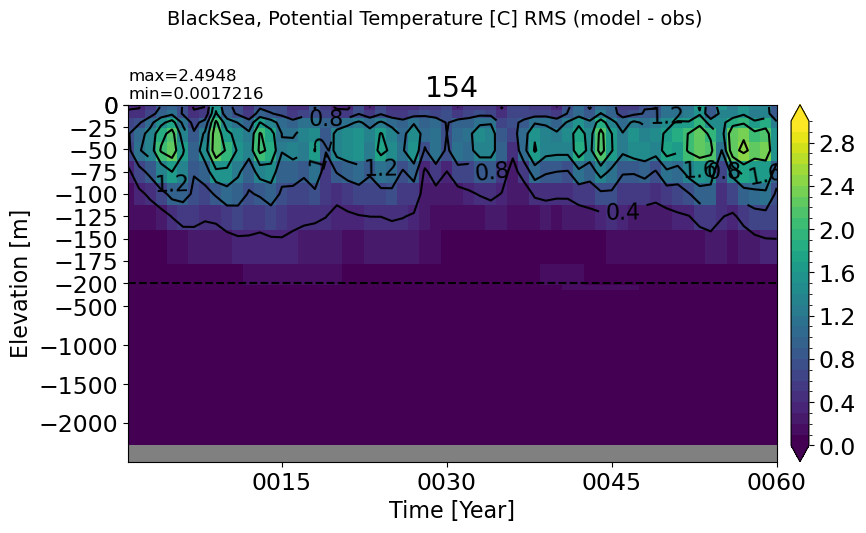
BlackSea#
make_temp_rmse_plot(m='BlackSea', splitscale = [0., -200., -2500])
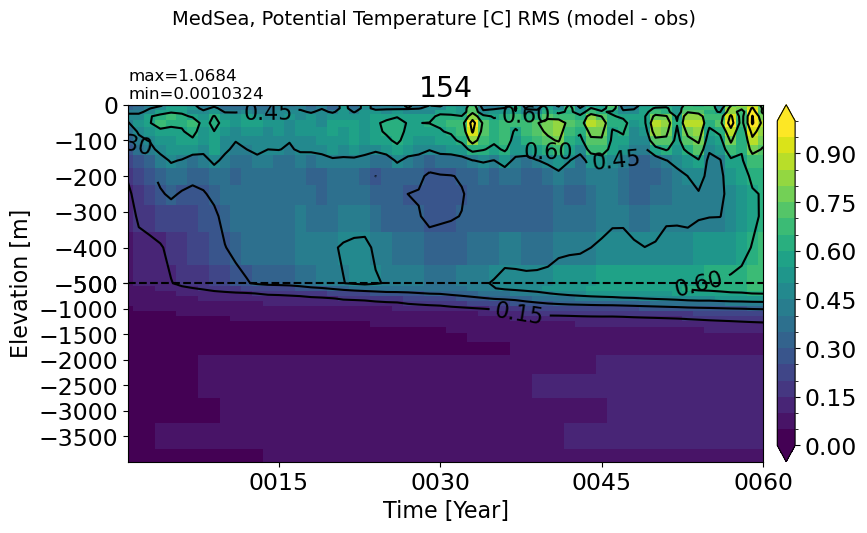
MedSea#
make_temp_rmse_plot(m='MedSea', vmax=1)
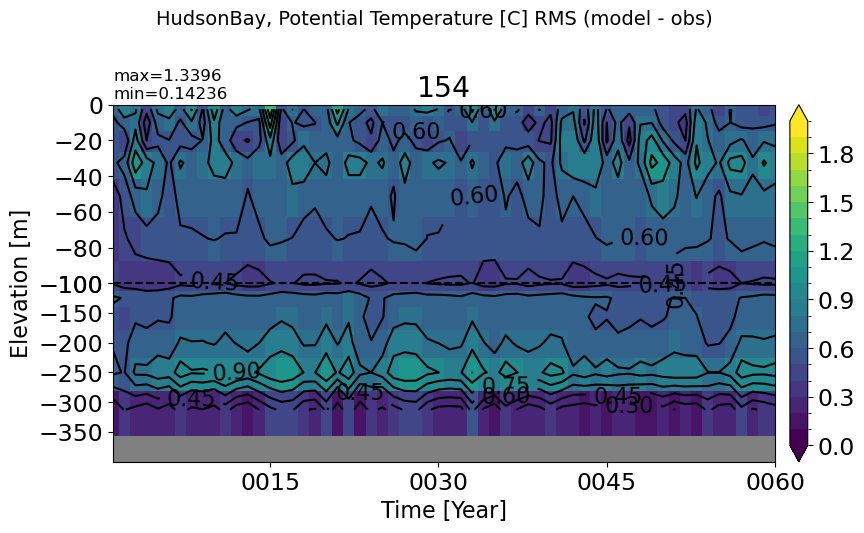
HudsonBay#
make_temp_rmse_plot(m='HudsonBay', vmax=2, splitscale = [0., -100., -400])
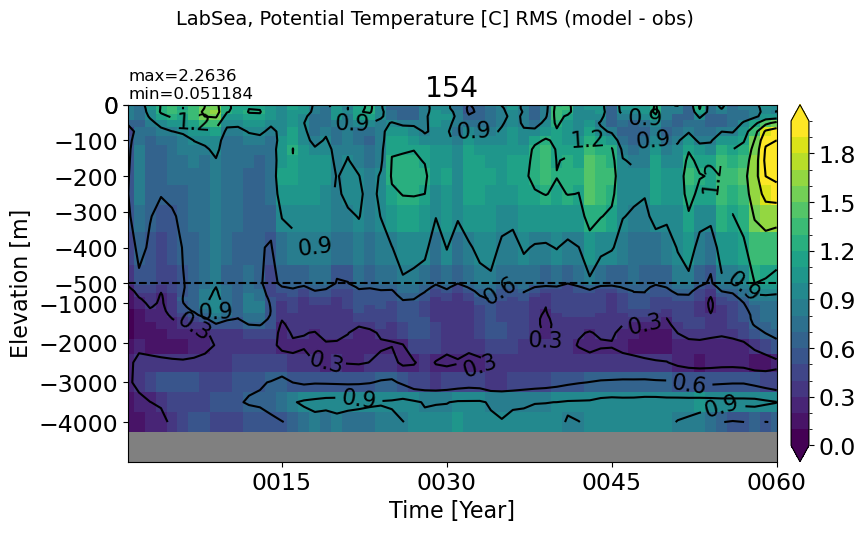
LabSea#
make_temp_rmse_plot(m='LabSea', vmax=2, splitscale = [0., -500., -5000])
BaffinBay#
make_temp_rmse_plot(m='BaffinBay', vmax=2, splitscale = [0., -500., -2400])
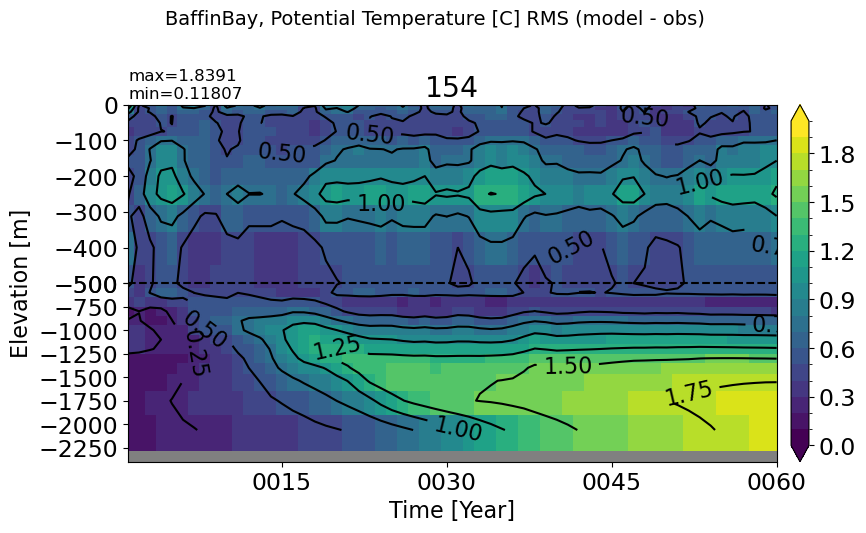
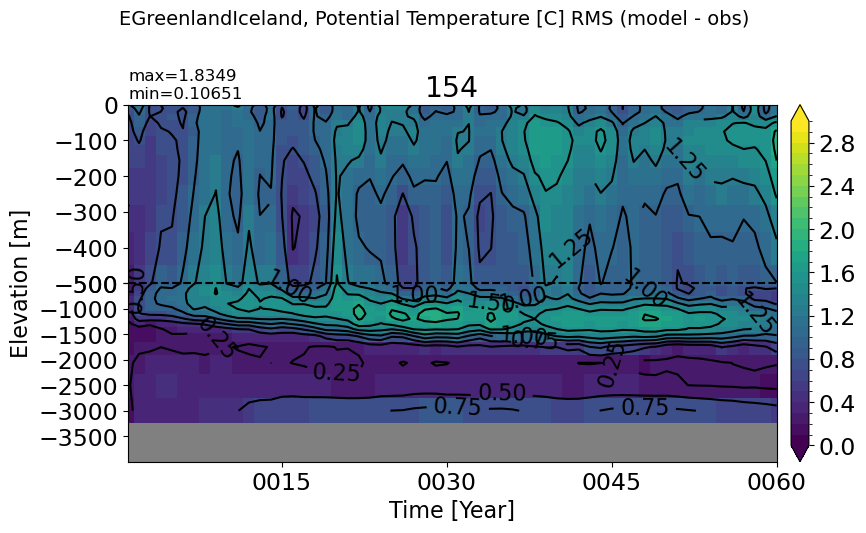
EGreenlandIceland#
make_temp_rmse_plot(m='EGreenlandIceland')
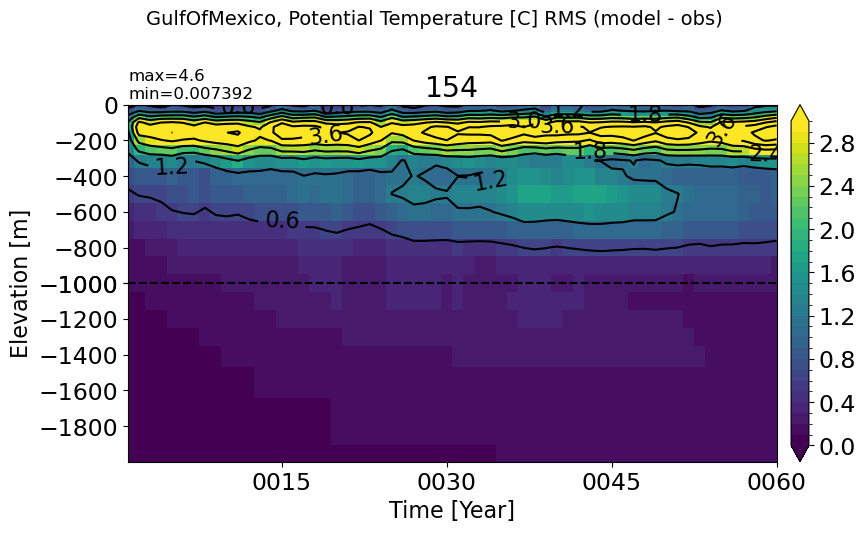
GulfOfMexico#
make_temp_rmse_plot(m='GulfOfMexico', splitscale = [0., -1000., -2000])
Salinity#
def make_salt_rmse_plot(m='Global', vmax=0.5, splitscale = [0., -500., -4000]):
vmin=0
for path, case, i in zip(ocn_path, casename, range(len(casename))):
fig, ax = plt.subplots(nrows=nr, ncols=nc, figsize=fs, sharex=False, sharey=True)
plt.suptitle(str(m)+', Salinity [psu] RMS (model - obs)', fontsize=14)
ds_mom = xr.open_dataset(path+case+'_so_rmse.nc').sel(time=slice('0001-01-01', end_date))
dummy_mom = np.ma.masked_invalid(ds_mom.sel(region=m).so_rms.values)
ztplot(dummy_mom, ds_mom.time.values, ds_mom.z_l.values*-1, ignore=np.nan, splitscale=splitscale,
contour=True, axis=ax , title=label[i], extend='both', colormap='viridis',
autocenter=True, tunits='Year', show=False, clim=(vmin, vmax))
plt.subplots_adjust(top = 0.8)
plt.tight_layout()
return
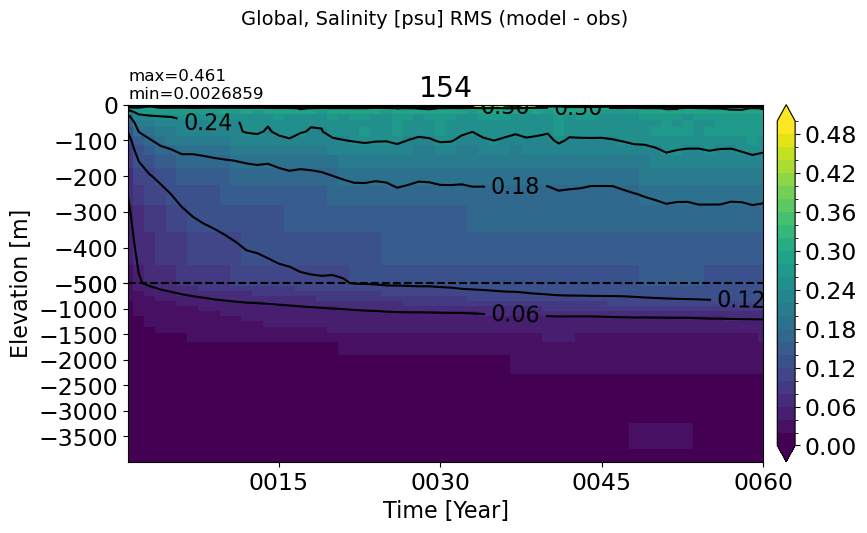
Global#
make_salt_rmse_plot()
Arctic#
make_salt_rmse_plot(m='Arctic', vmax=1)
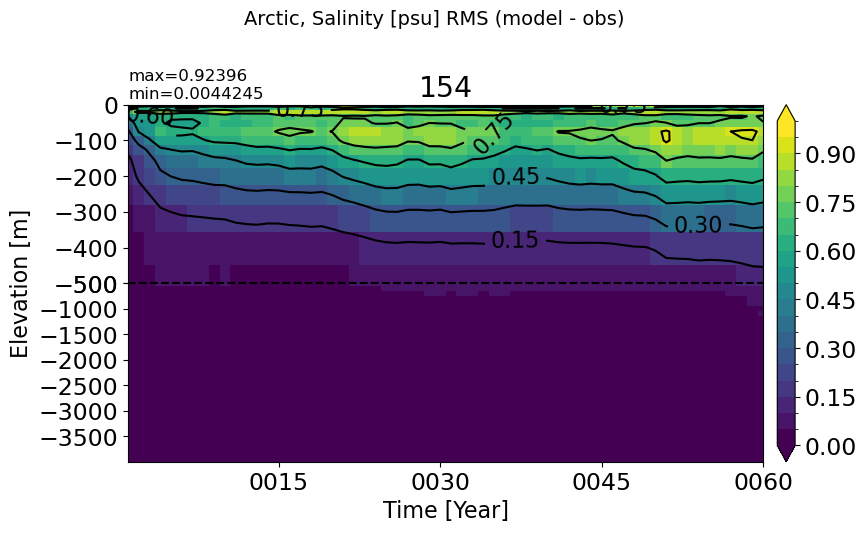
SouthernOcean#
make_salt_rmse_plot(m='SouthernOcean', vmax=0.3)
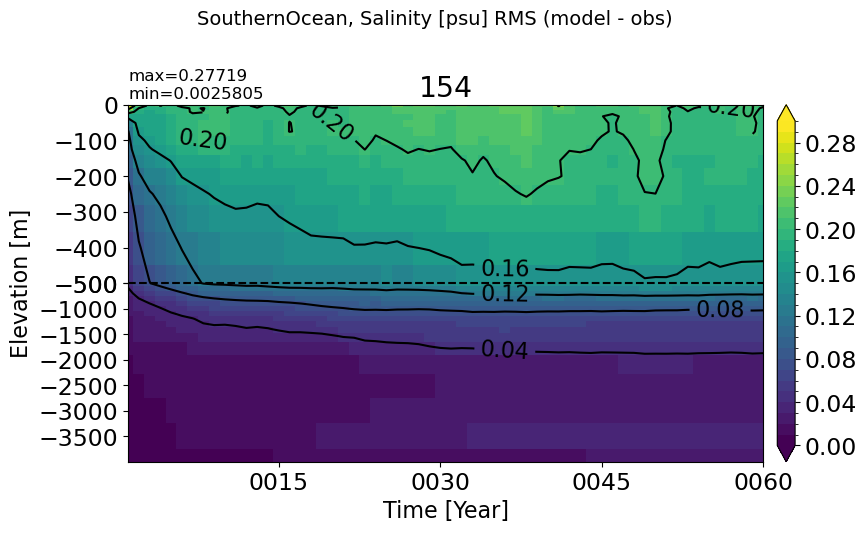
SouthernOcean60S#
make_salt_rmse_plot(m='SouthernOcean60S', vmax=0.3)
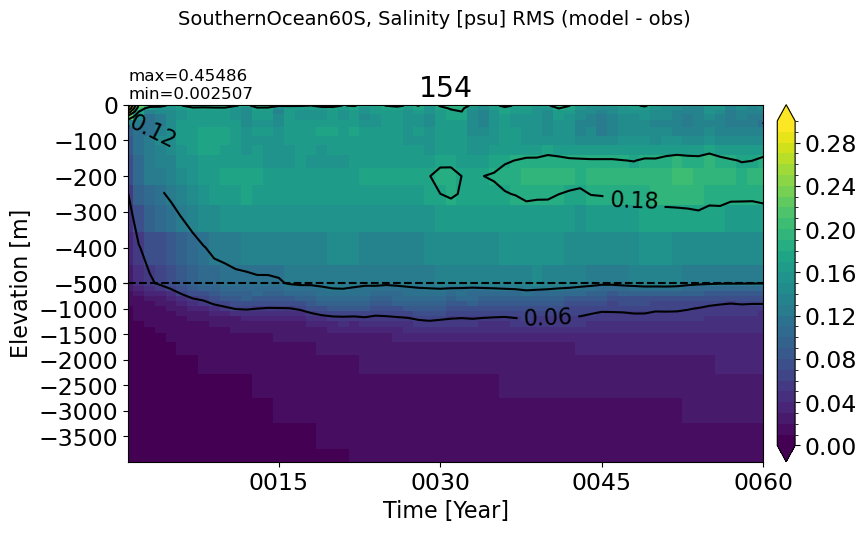
PacificOcean#
make_salt_rmse_plot(m='PacificOcean', vmax=0.3)
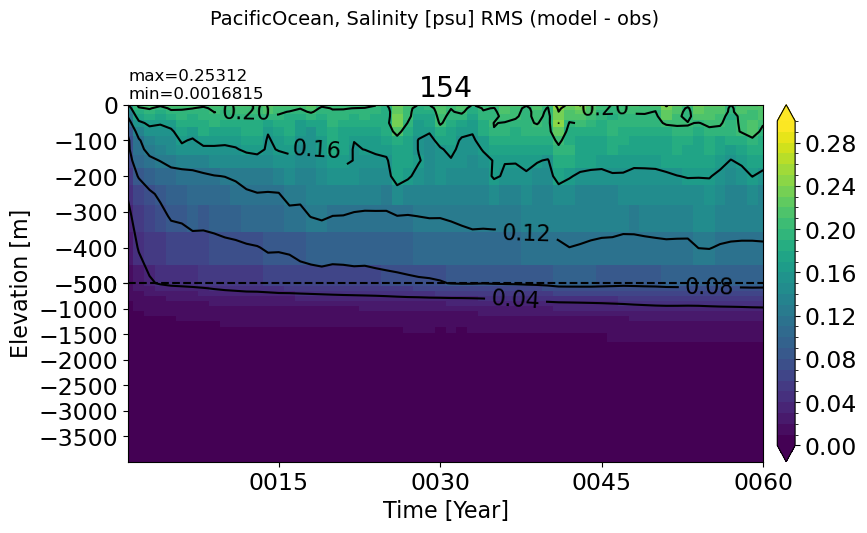
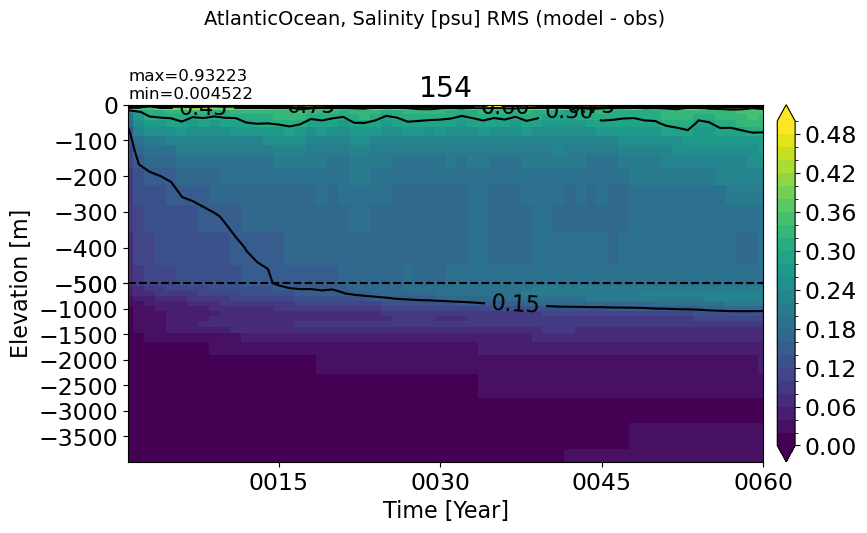
AtlanticOcean#
make_salt_rmse_plot(m='AtlanticOcean')
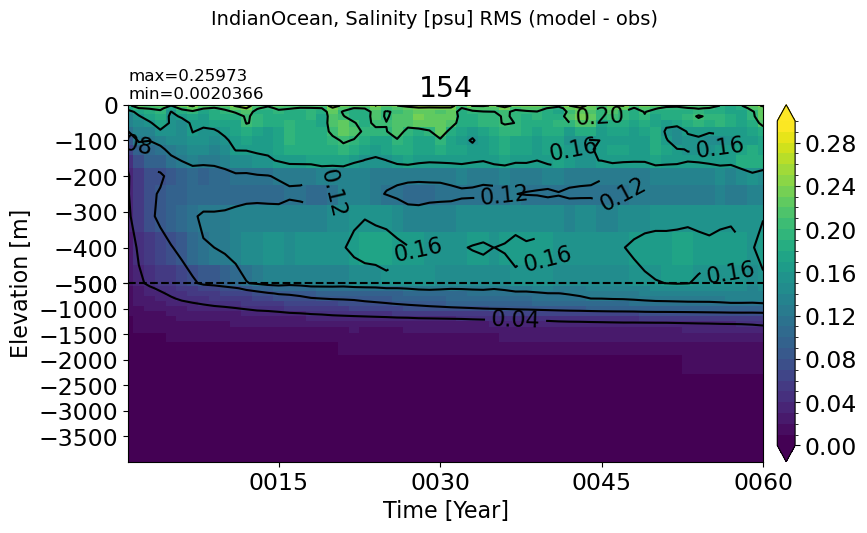
IndianOcean#
make_salt_rmse_plot(m='IndianOcean', vmax=0.3)
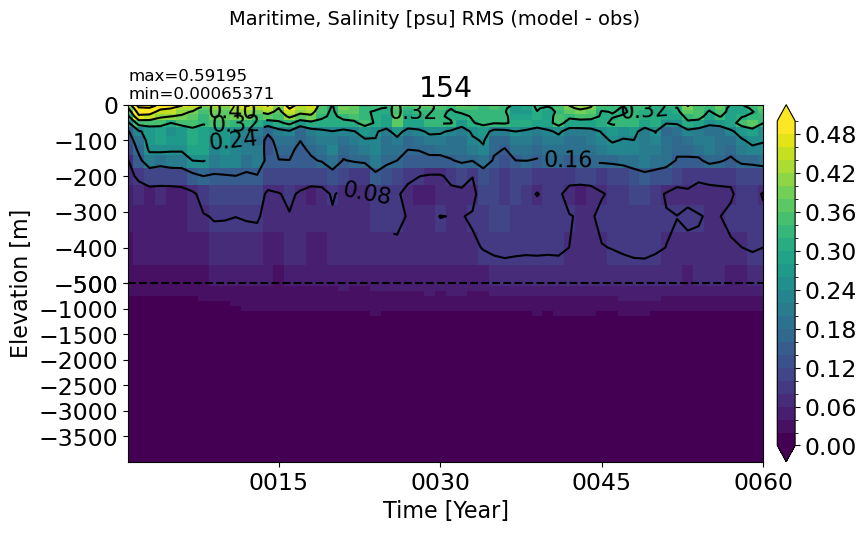
Maritime#
make_salt_rmse_plot(m='Maritime')
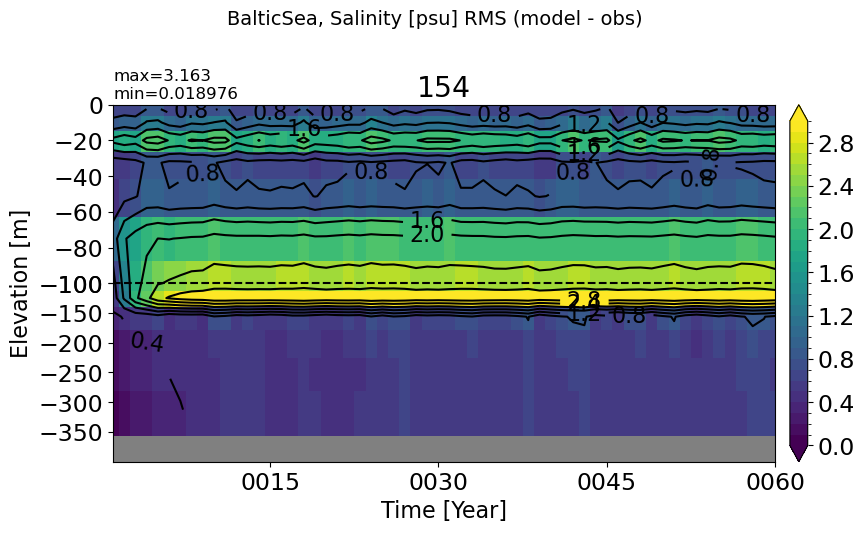
BalticSea#
make_salt_rmse_plot(m='BalticSea', vmax=3, splitscale = [0., -100., -400])
PersianGulf#
make_salt_rmse_plot(m='PersianGulf', vmax=1.5, splitscale = [0., -100., -70])
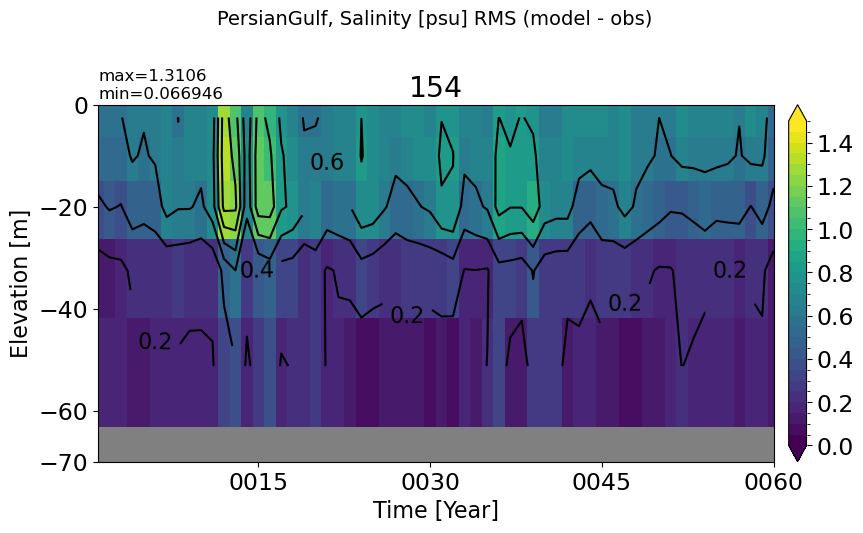
RedSea#
make_salt_rmse_plot(m='RedSea', vmax=1.0, splitscale = [0., -200., -1200])
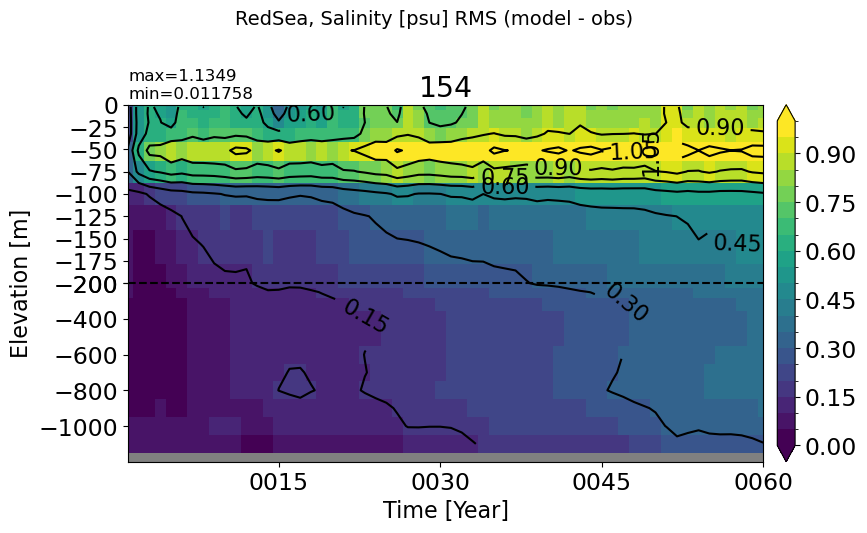
BlackSea#
make_salt_rmse_plot(m='BlackSea', vmax=1.5, splitscale = [0., -200., -2500])
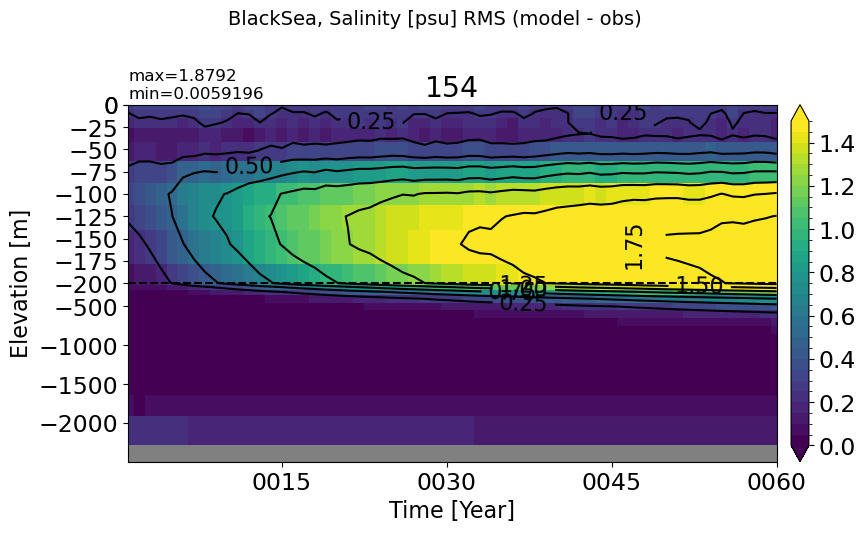
MedSea#
make_salt_rmse_plot(m='MedSea')
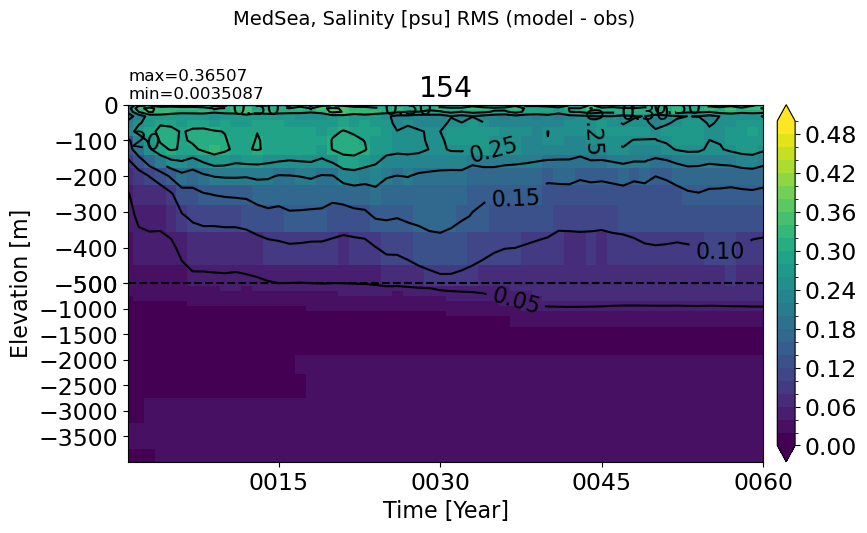
HudsonBay#
make_salt_rmse_plot(m='HudsonBay', vmax=1.5, splitscale = [0., -100., -400])
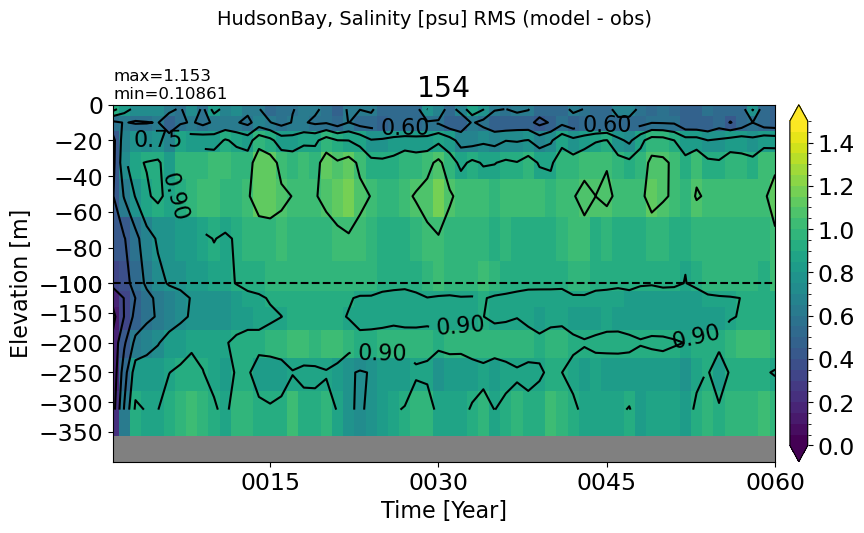
LabSea#
make_salt_rmse_plot(m='LabSea', vmax=0.5, splitscale = [0., -500., -5000])
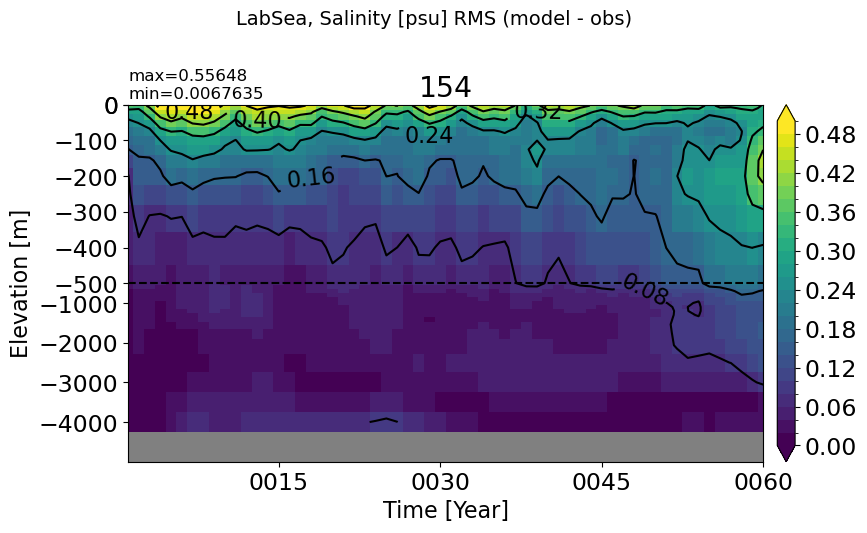
BaffinBay#
make_salt_rmse_plot(m='BaffinBay', vmax=0.5, splitscale = [0., -500., -2400])
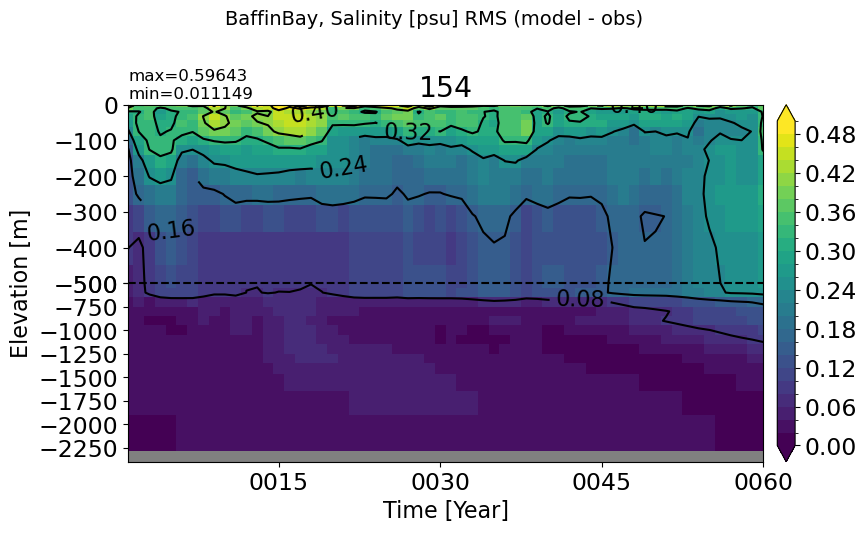
EGreenlandIceland#
make_salt_rmse_plot(m='EGreenlandIceland')
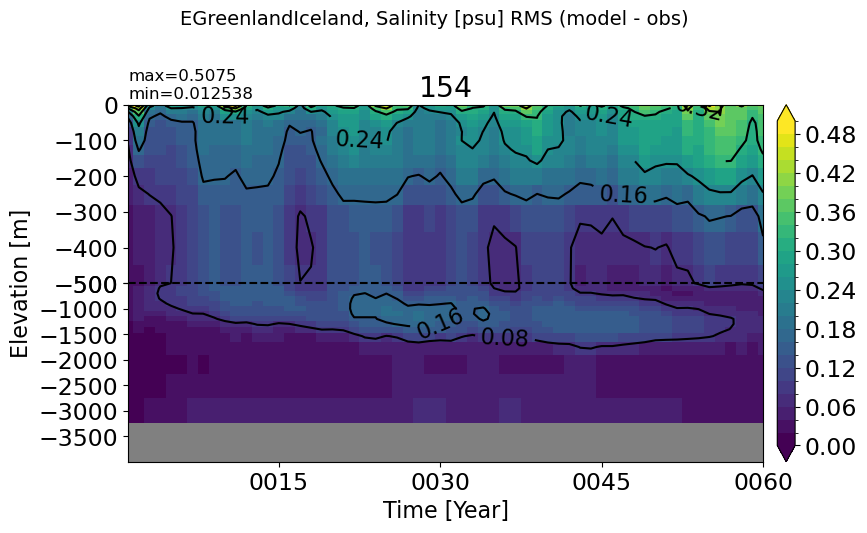
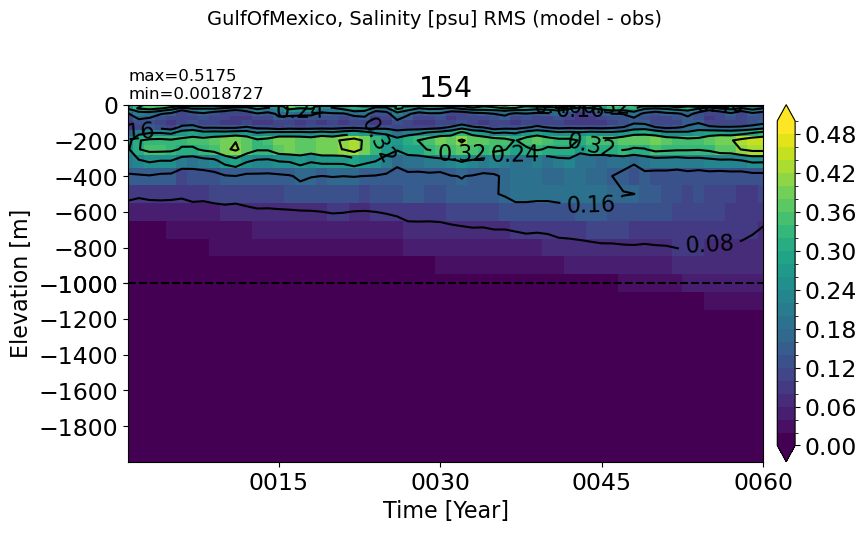
GulfOfMexico#
make_salt_rmse_plot(m='GulfOfMexico', splitscale = [0., -1000., -2000])
Biases at selected vertical levels#
Potential Temperature#
def make_temp_level_plot(k=0, t=5):
for path, case, i in zip(ocn_path, casename, range(len(casename))):
ds_mom_t = xr.open_dataset(path+case+'_thetao_time_mean.nc')
if 'area_t' in dir(grd[i]):
area = grd[i].area_t
else:
area = grd[i].areacello
fig, ax = plt.subplots(nrows=1, ncols=1, figsize=(12,6.5))
temp_mom = np.ma.masked_invalid(ds_mom_t.thetao_bias[k,:].values)
plt.suptitle('Temperature bias [C] at depth = {} m (level = {})'.format(ds_mom_t.z_l[k].values, k))
xyplot(temp_mom, grd[i].geolon, grd[i].geolat, area, title=str(label[i]+' - obs'), axis=ax,
clim=(-t,t), nbins=100, colormap=plt.cm.bwr, centerlabels=True)
plt.subplots_adjust(top = 0.8)
return
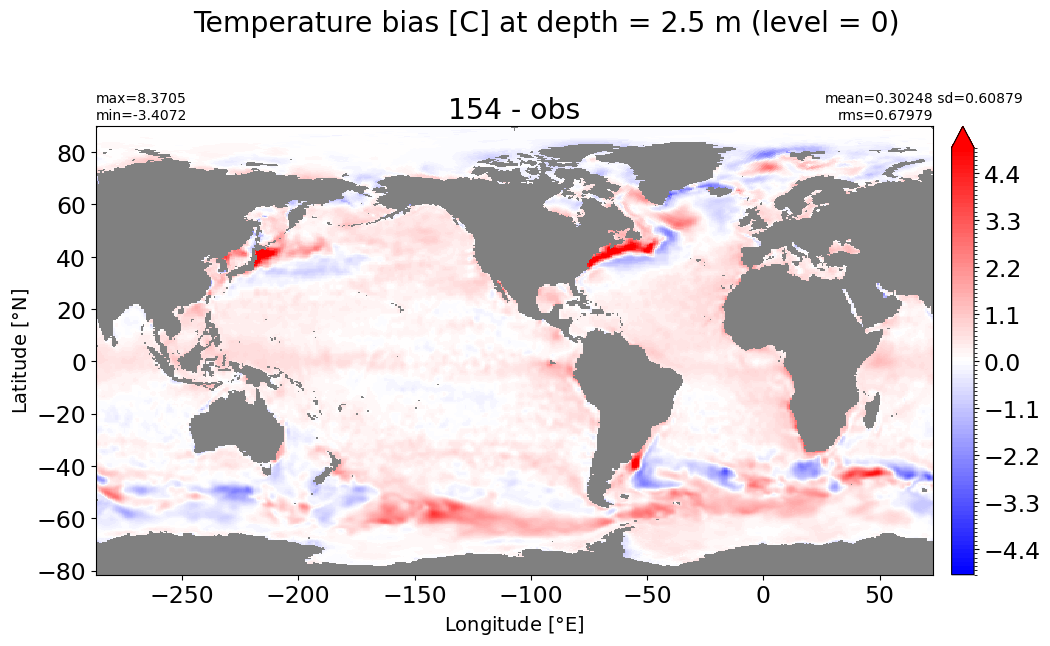
Depth = 2.5 m, k=0#
make_temp_level_plot(k=0, t=5)
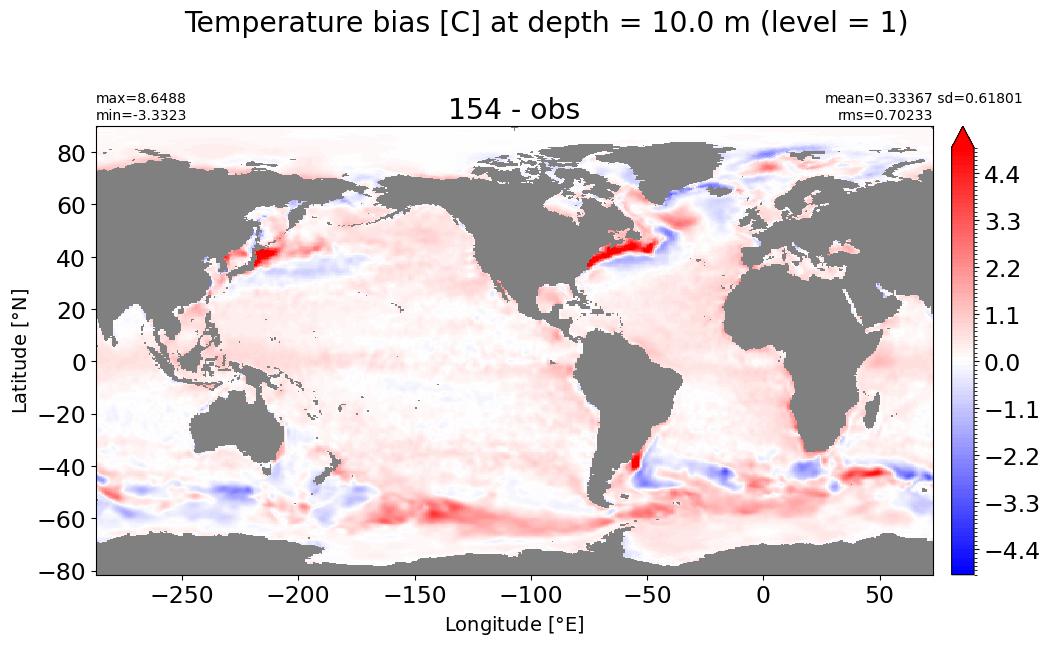
Depth = 10 m, k=1#
make_temp_level_plot(k=1, t=5)
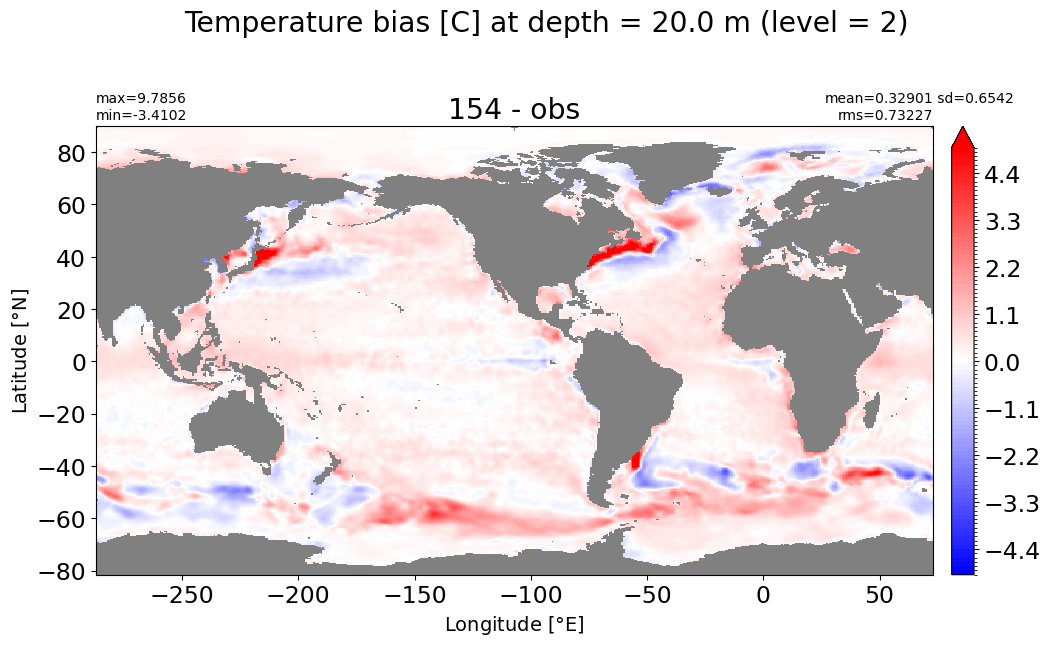
Depth = 20 m, k=2#
make_temp_level_plot(k=2, t=5)
Depth = 32.5 m, k=3#
make_temp_level_plot(k=3, t=5)
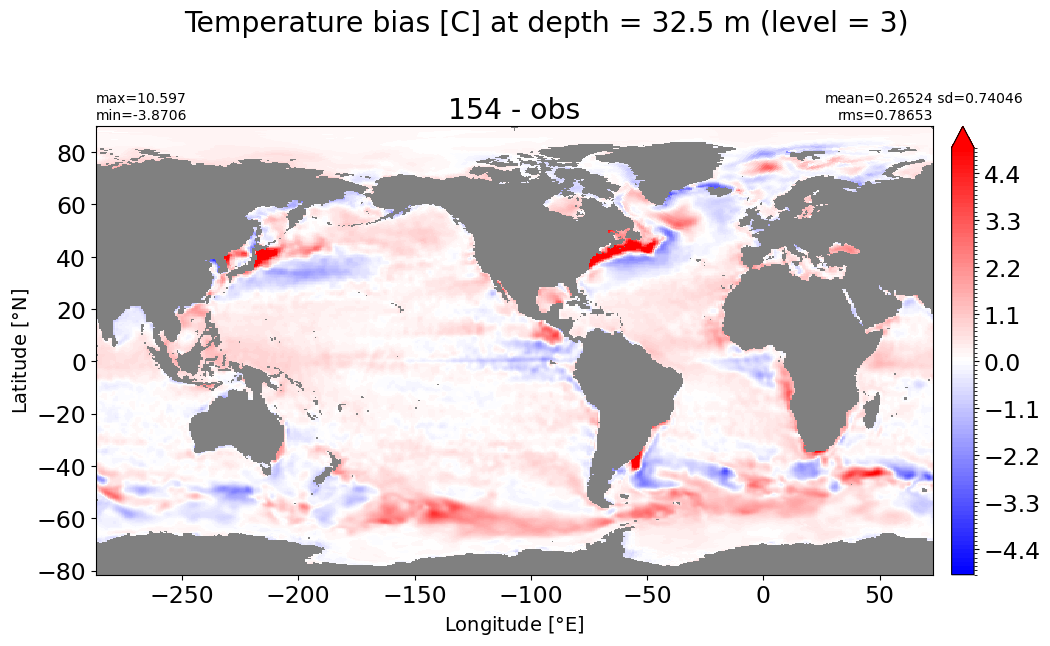
Depth = 51.25 m, k=4#
make_temp_level_plot(k=4, t=8)
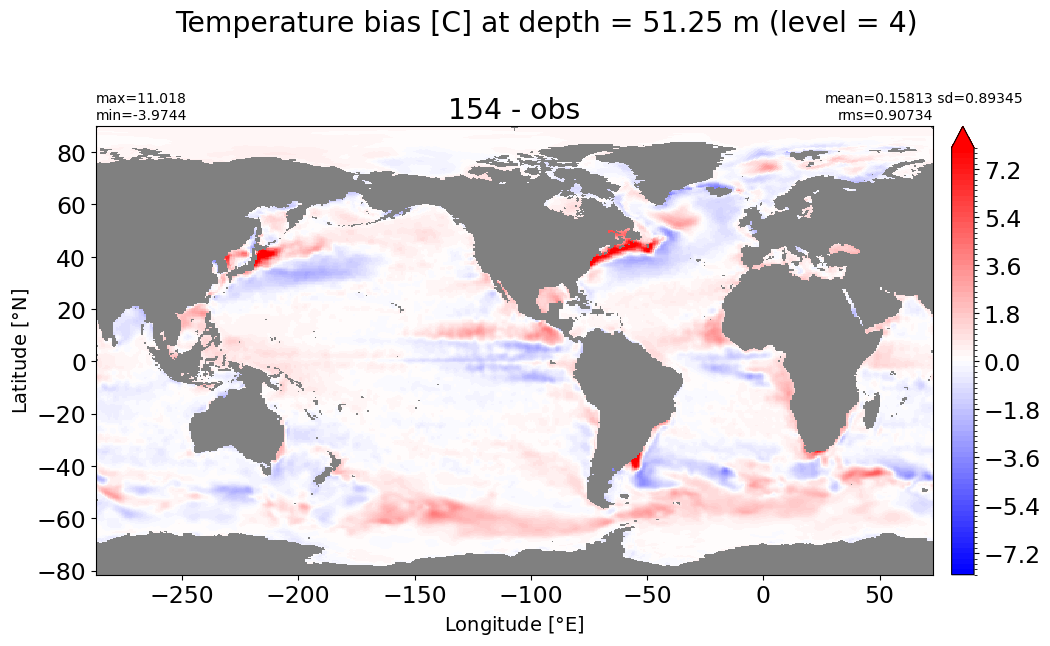
Depth = 75 m, k=5#
make_temp_level_plot(k=5, t=10)
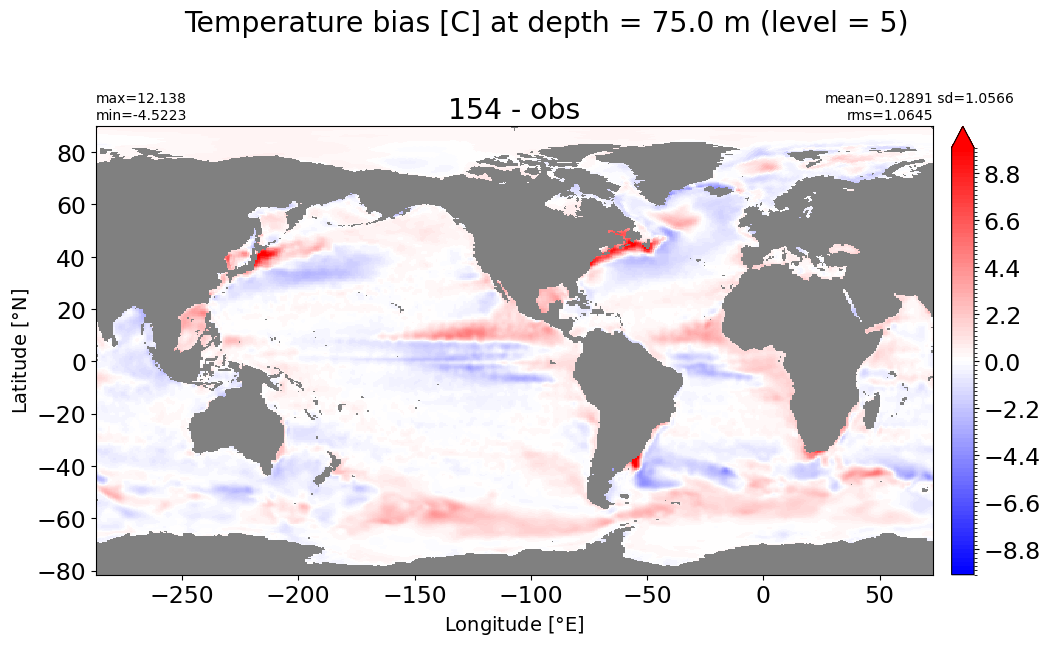
Depth = 100 m, k=6#
make_temp_level_plot(k=6, t=10)
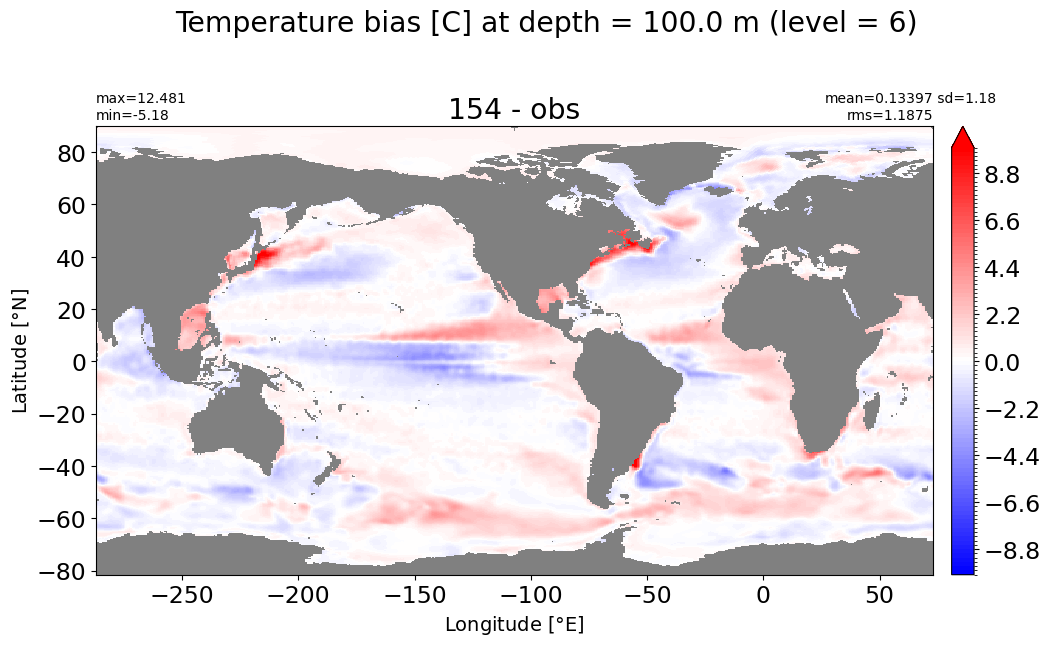
Depth = 125 m, k=7#
make_temp_level_plot(k=7, t=10)

Depth = 200 m, k=9#
make_temp_level_plot(k=9, t=8)
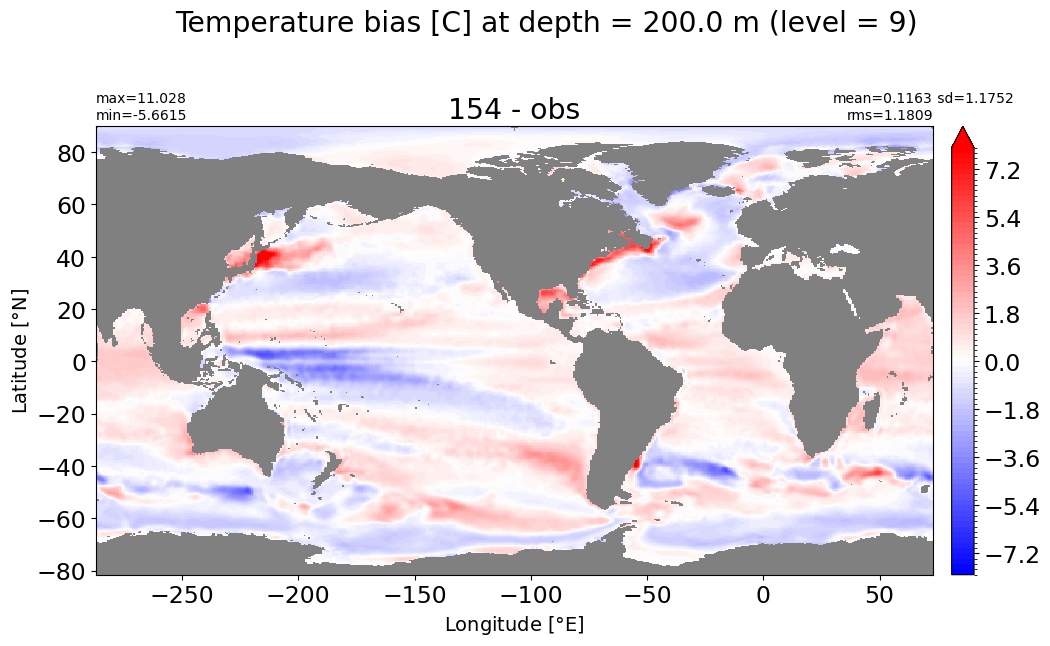
Depth = 250 m, k=10#
make_temp_level_plot(k=10, t=8)
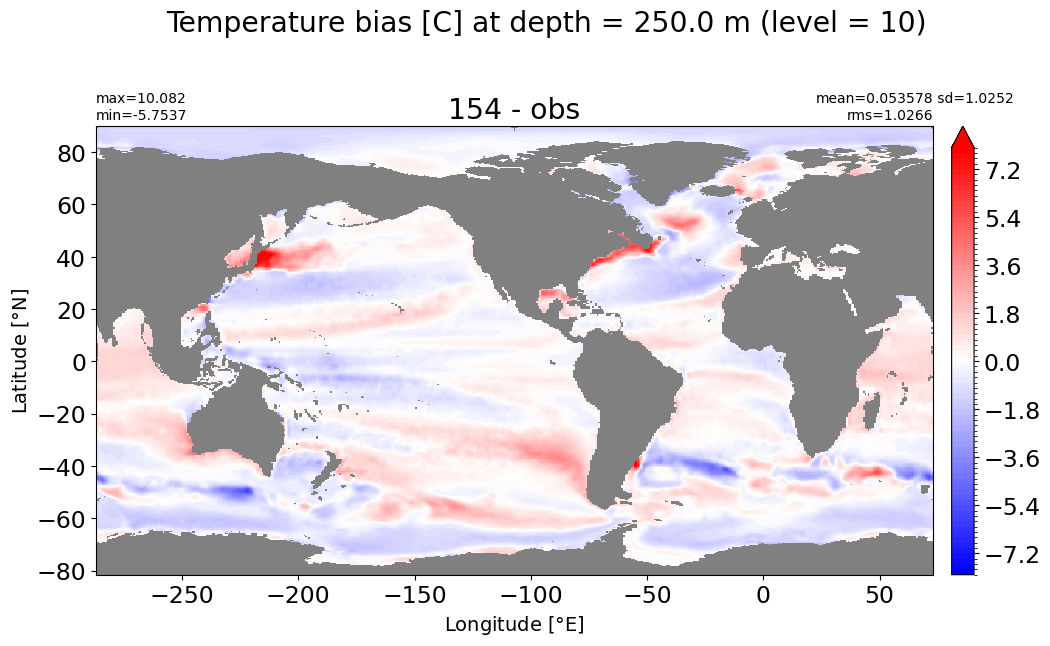
Depth = 312.25 m, k=11#
make_temp_level_plot(k=11, t=5)
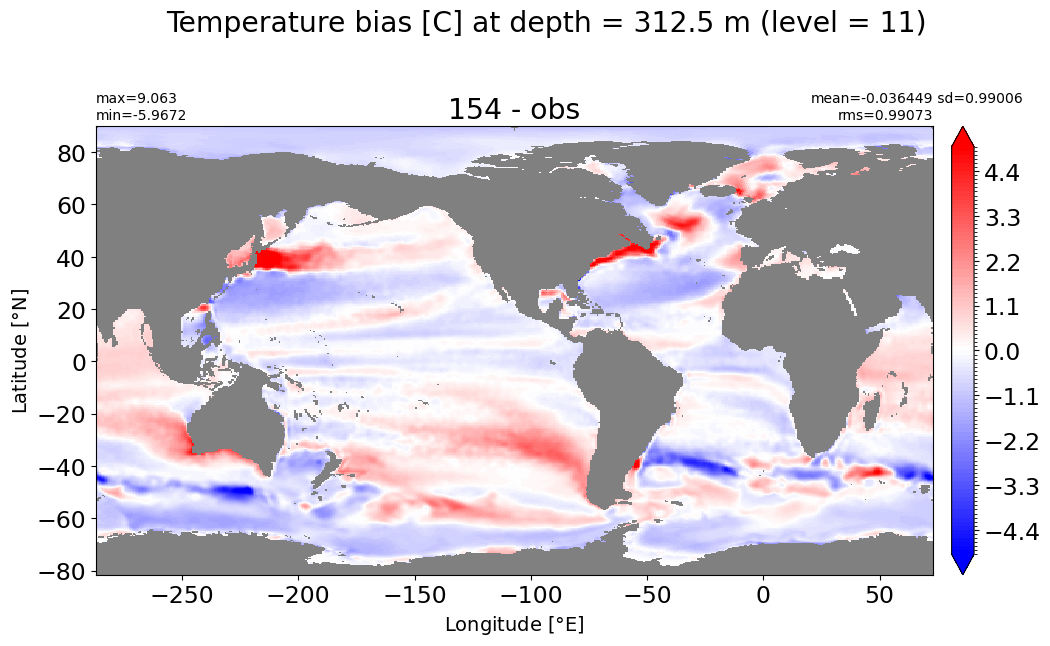
Depth = 400 m, k=12#
make_temp_level_plot(k=12, t=5)
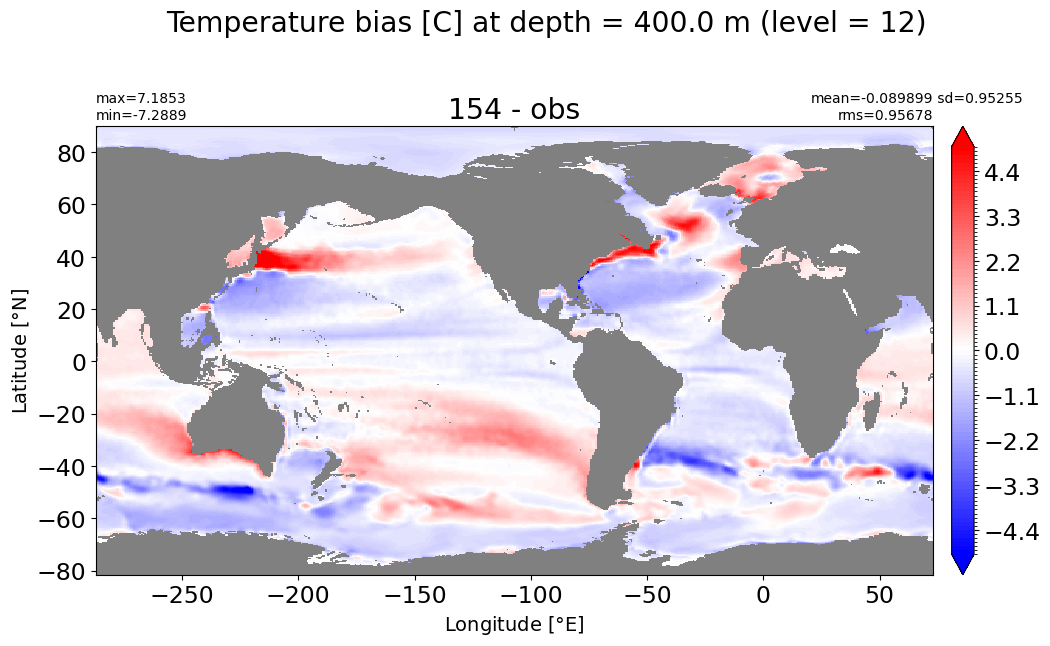
Depth = 500 m, k=13#
make_temp_level_plot(k=13, t=5)
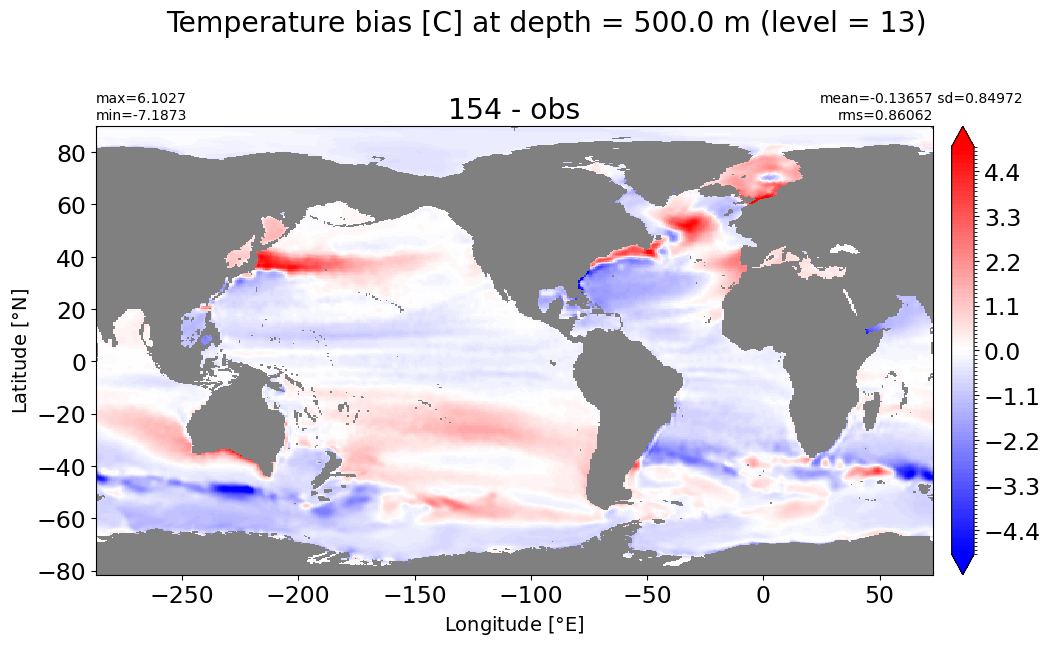
Depth = 600 m, k=14#
make_temp_level_plot(k=14, t=5)
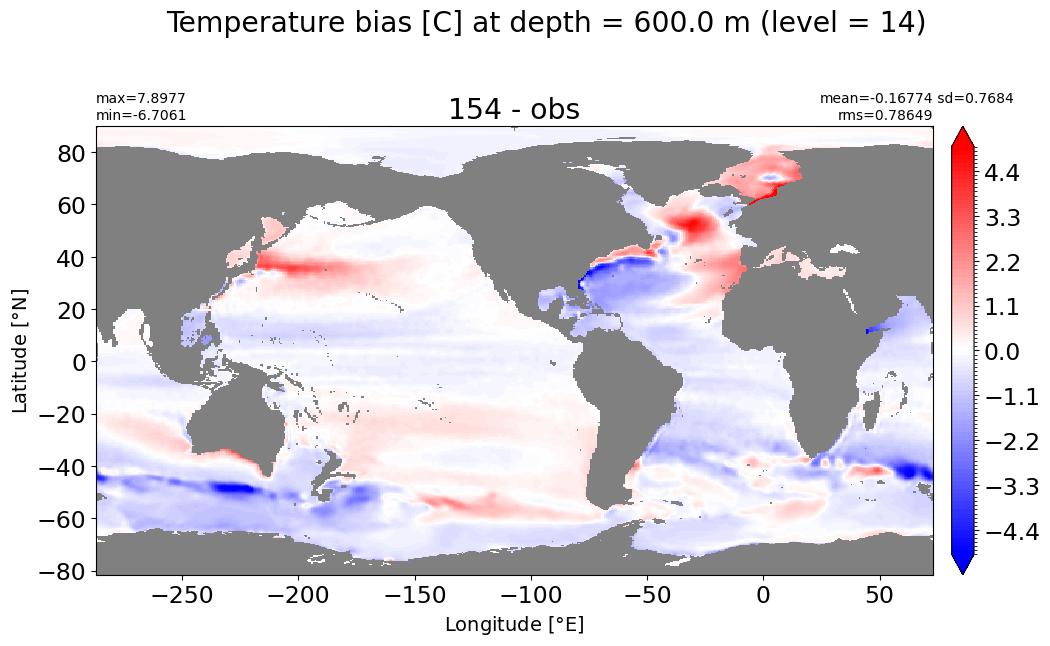
Depth = 700 m, k=15#
make_temp_level_plot(k=15, t=5)
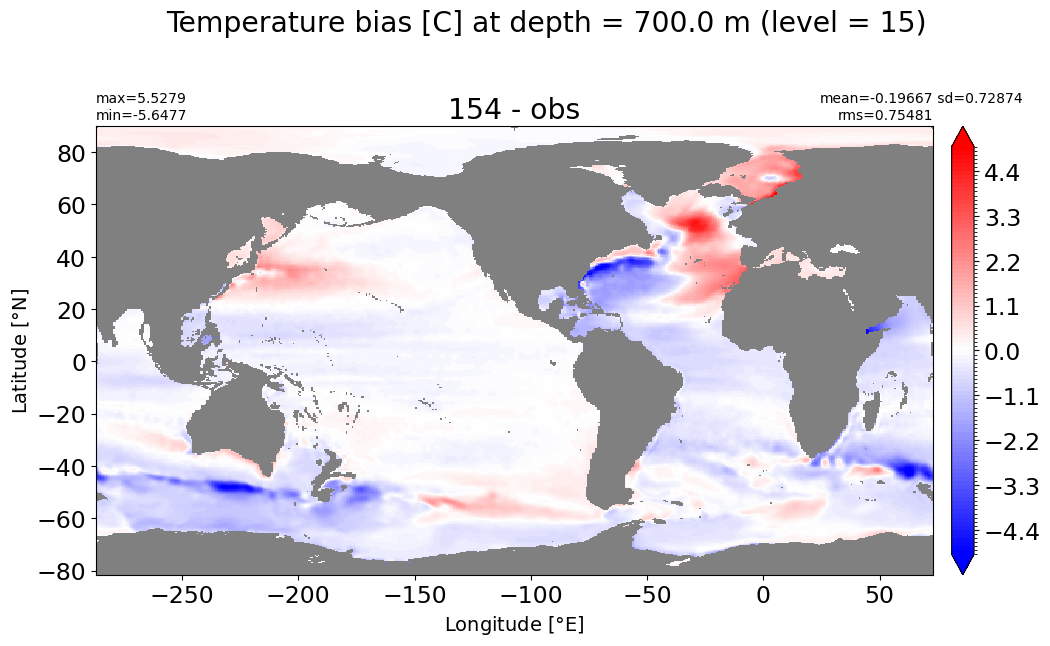
Depth = 800 m, k=16#
make_temp_level_plot(k=16, t=5)
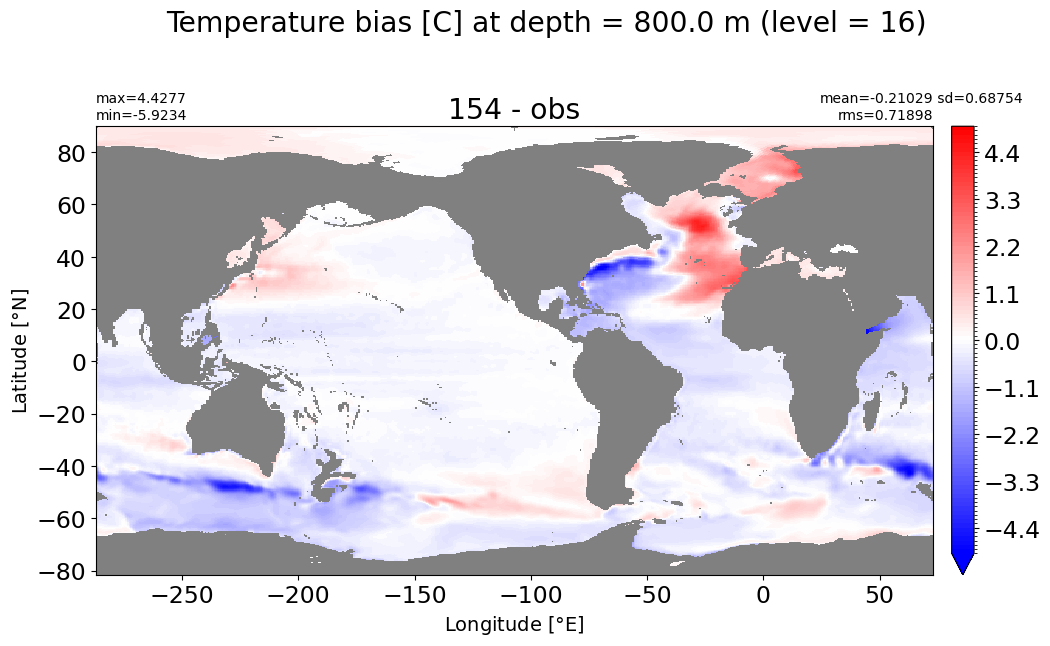
Depth = 900 m, k=17#
make_temp_level_plot(k=17, t=3)
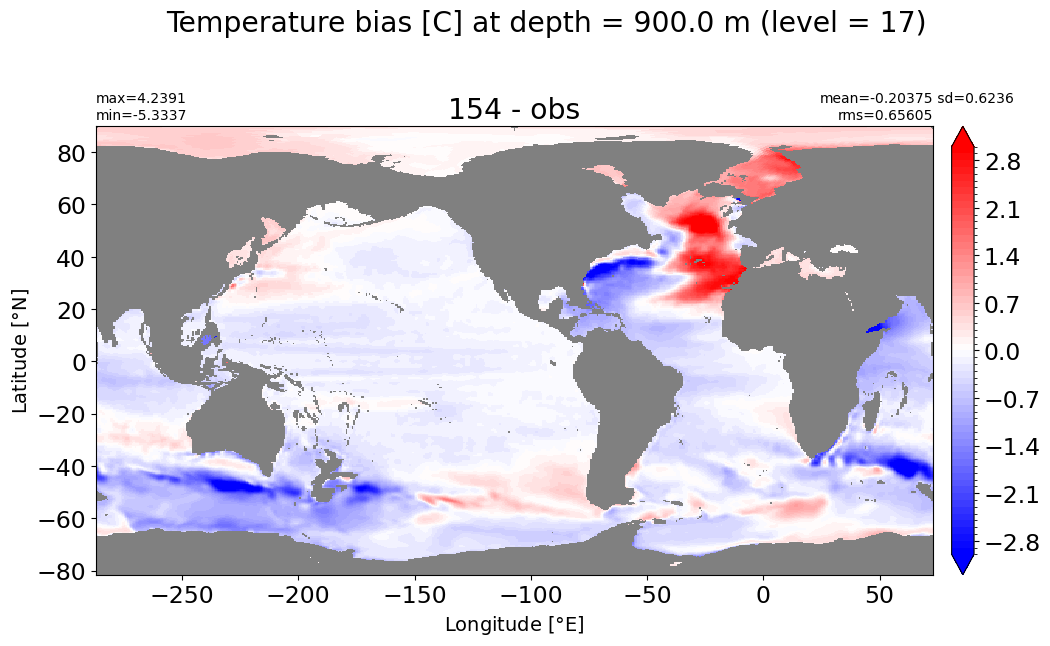
Depth = 1000 m, k=18#
make_temp_level_plot(k=18, t=3)
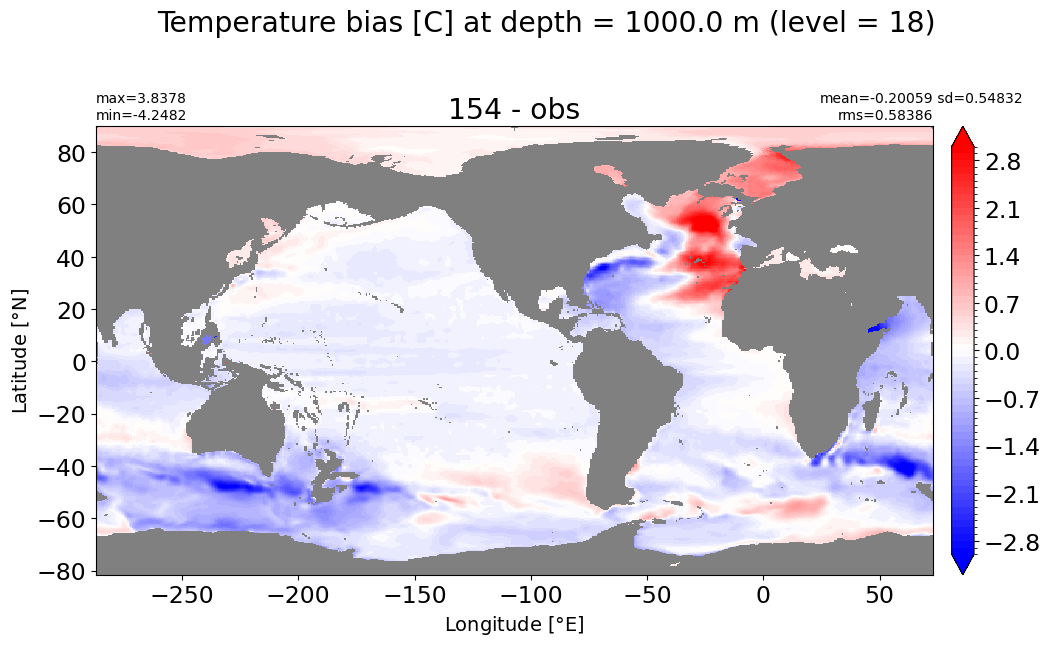
Depth = 1100 m, k=19#
make_temp_level_plot(k=19, t=3)
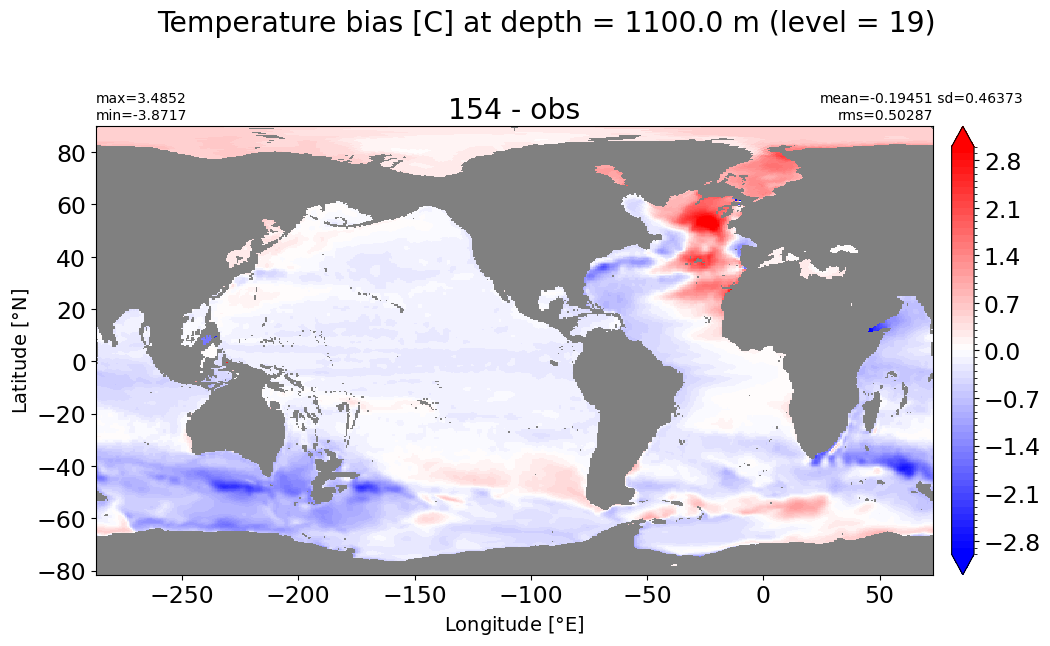
Depth = 1200 m, k=20#
make_temp_level_plot(k=20, t=2)
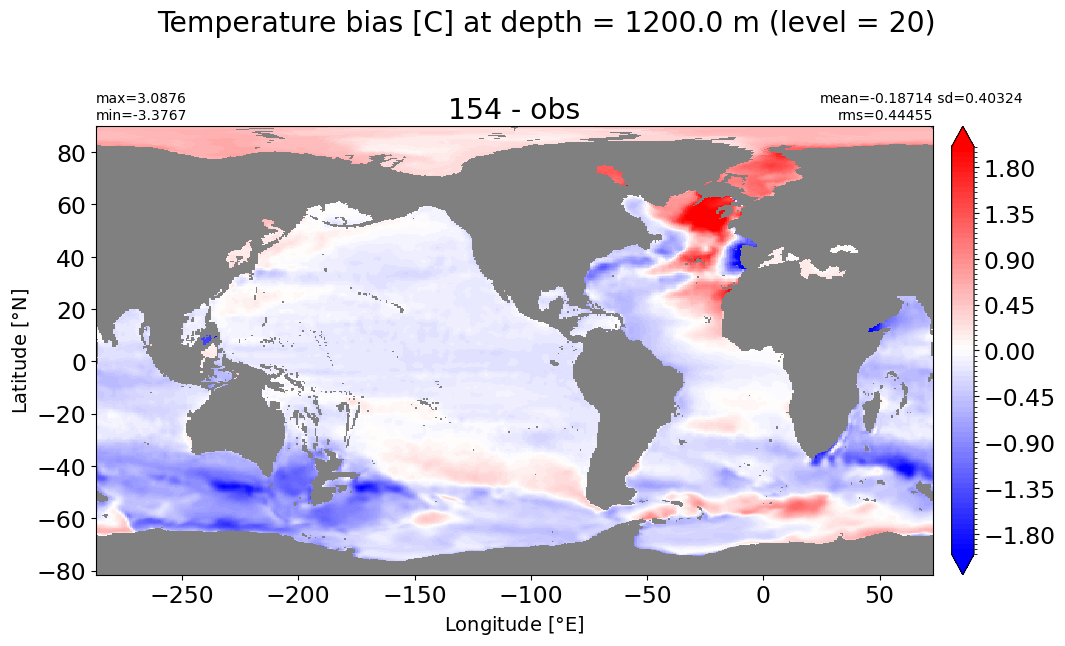
Depth = 1300 m, k=21#
make_temp_level_plot(k=21, t=2)
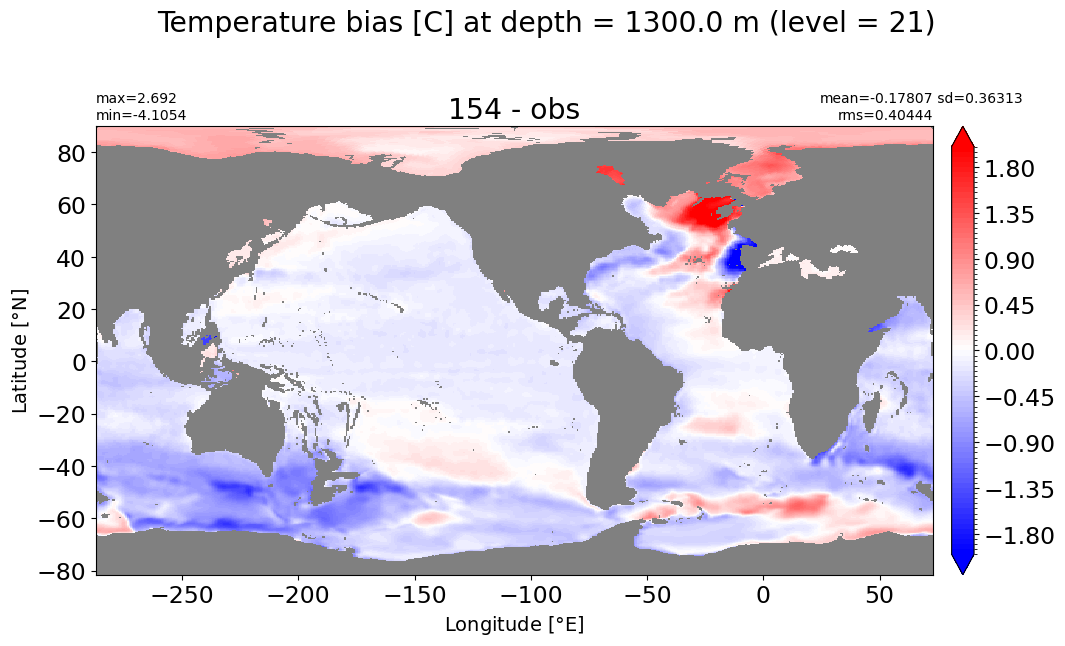
Depth = 1400 m, k=22#
make_temp_level_plot(k=22, t=2)
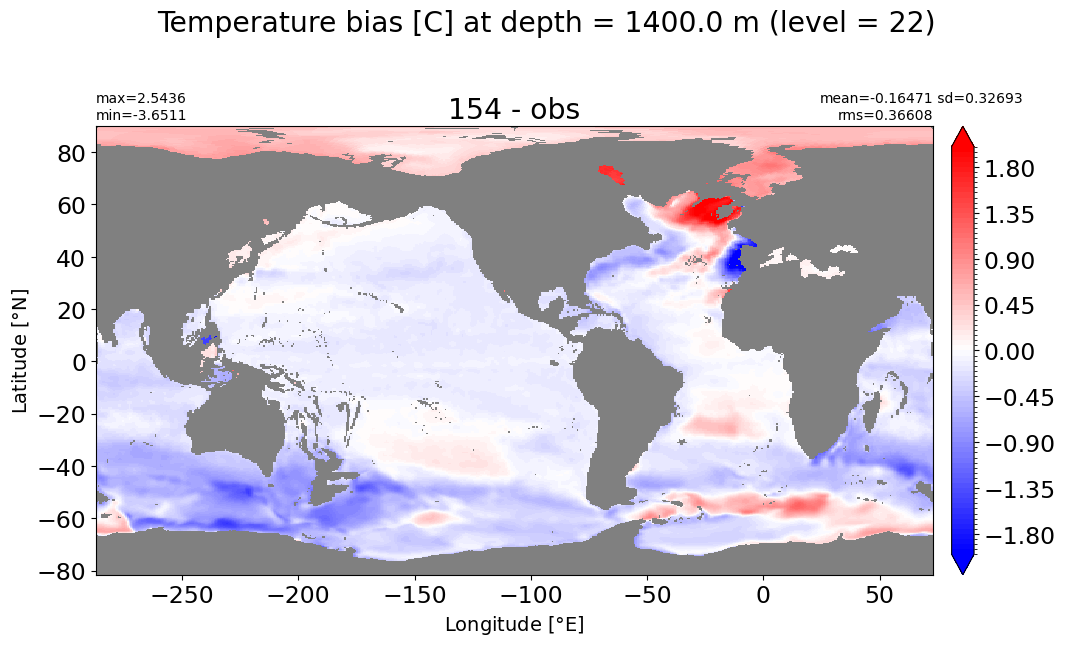
Depth = 1537.5 m, k=23#
make_temp_level_plot(k=23, t=2)
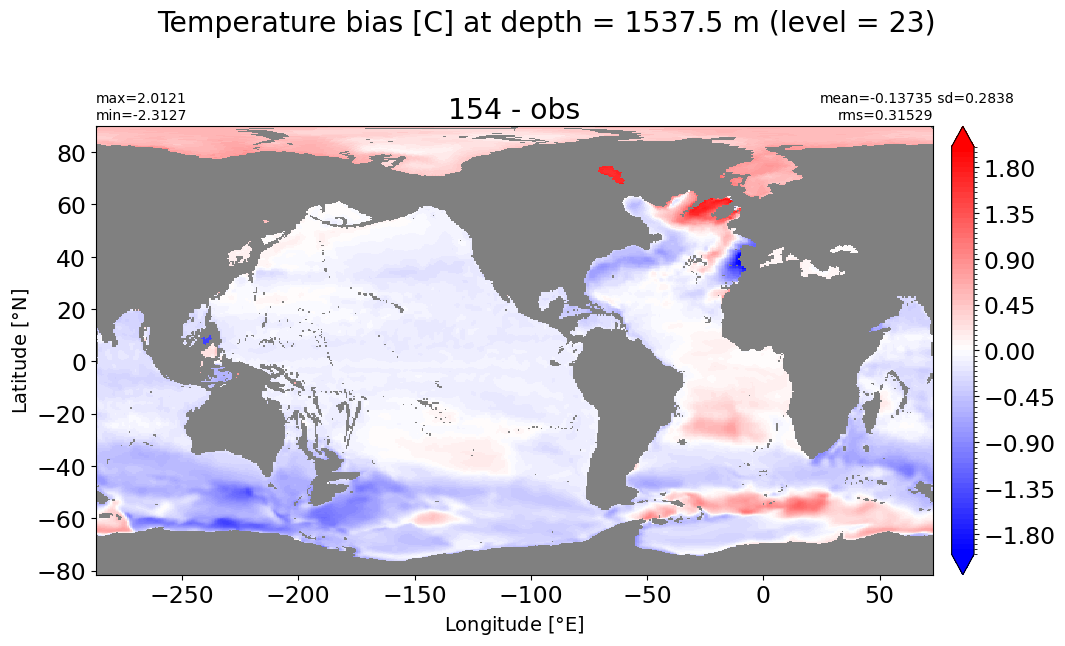
Depth = 1750 m, k=24#
make_temp_level_plot(k=24, t=1.5)
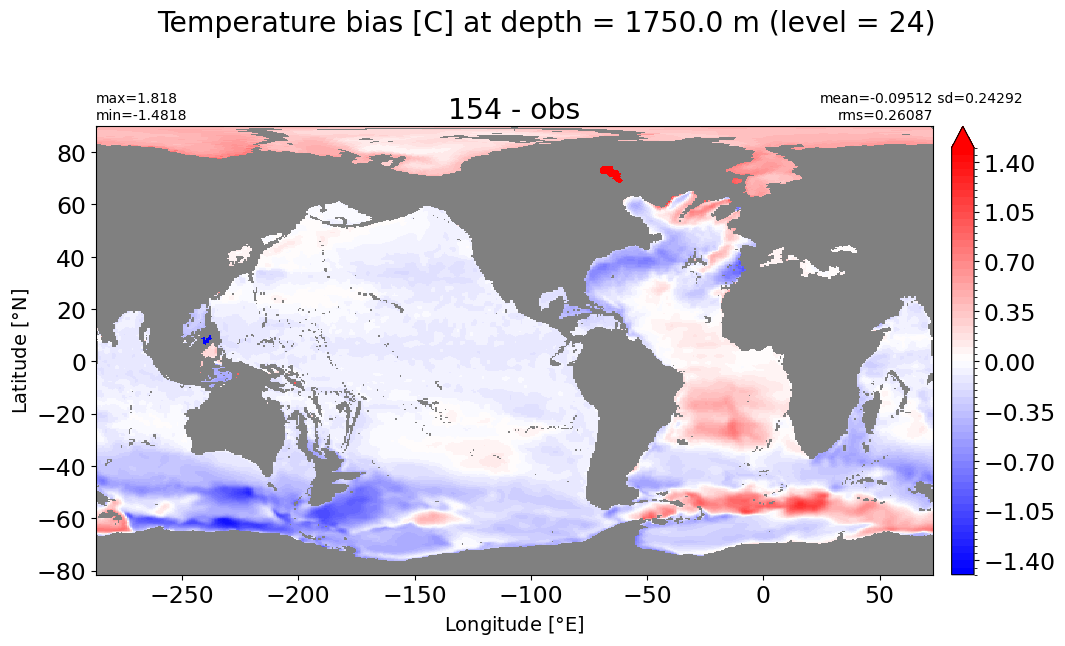
Depth = 2062.5 m, k=25#
make_temp_level_plot(k=25, t=1.5)
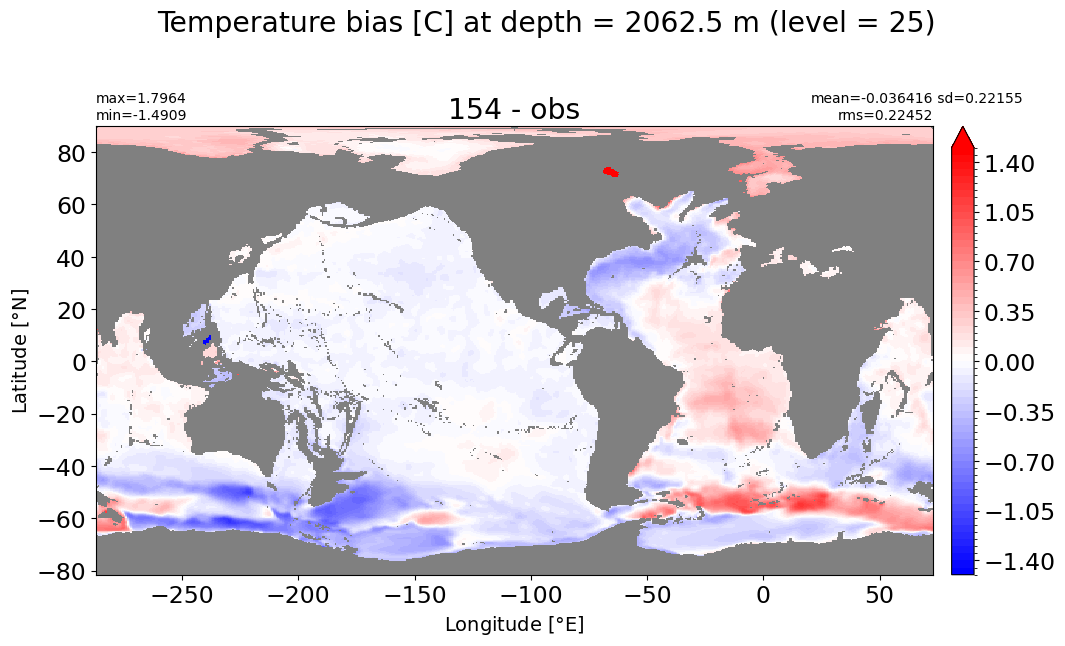
Depth = 2500 m, k=26#
make_temp_level_plot(k=26, t=1.5)
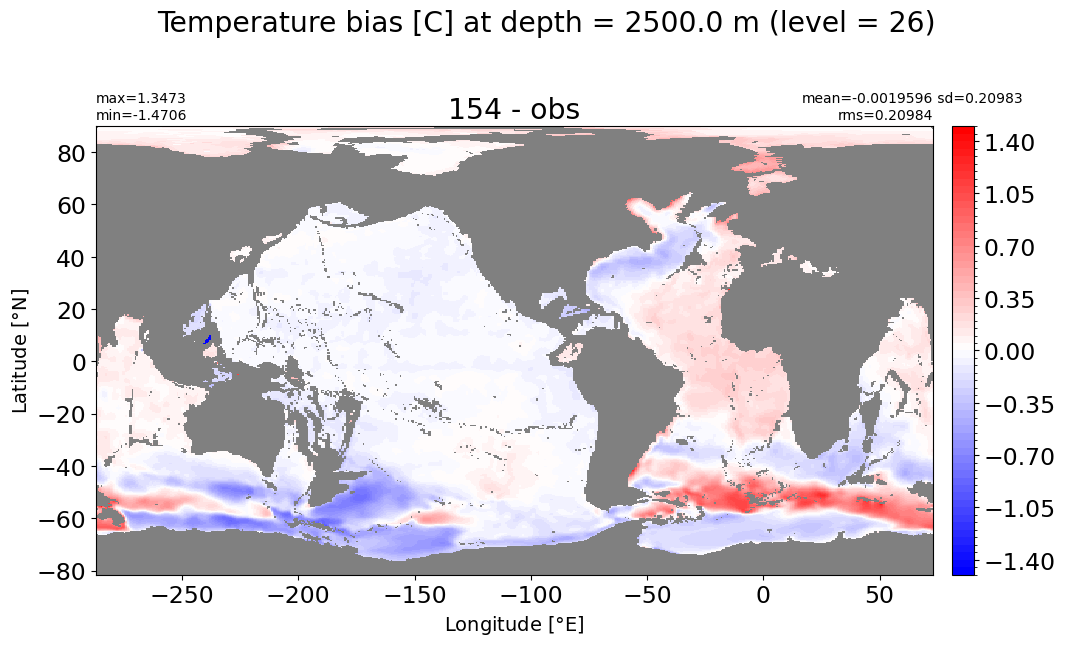
Depth = 3000 m, k=27#
make_temp_level_plot(k=27, t=1.)
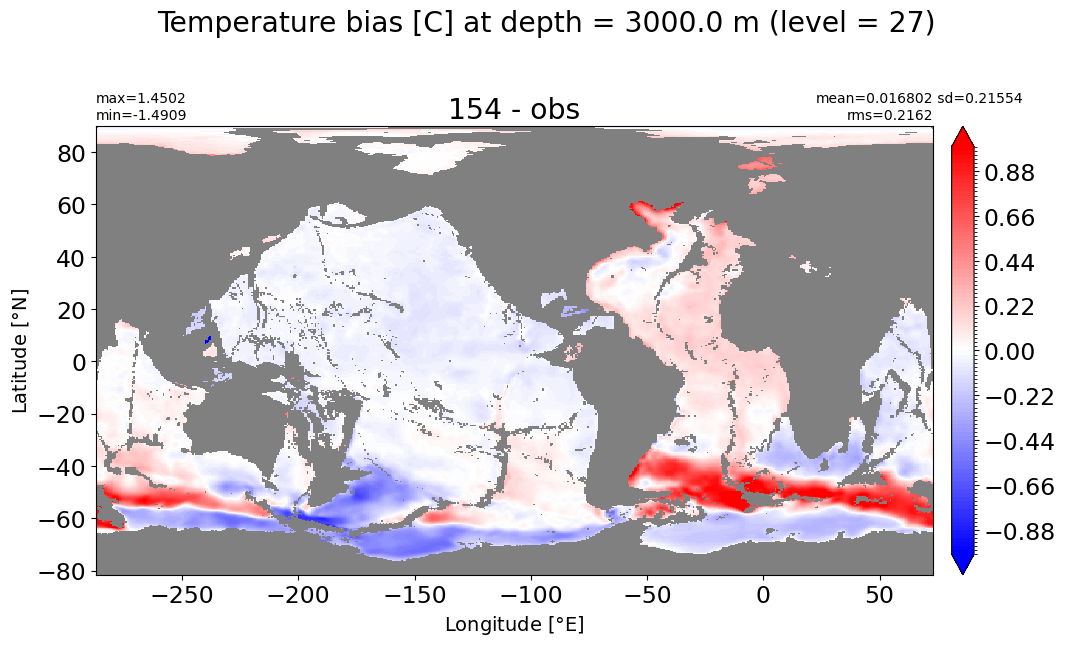
Depth = 3500 m, k=28#
make_temp_level_plot(k=28, t=1.)
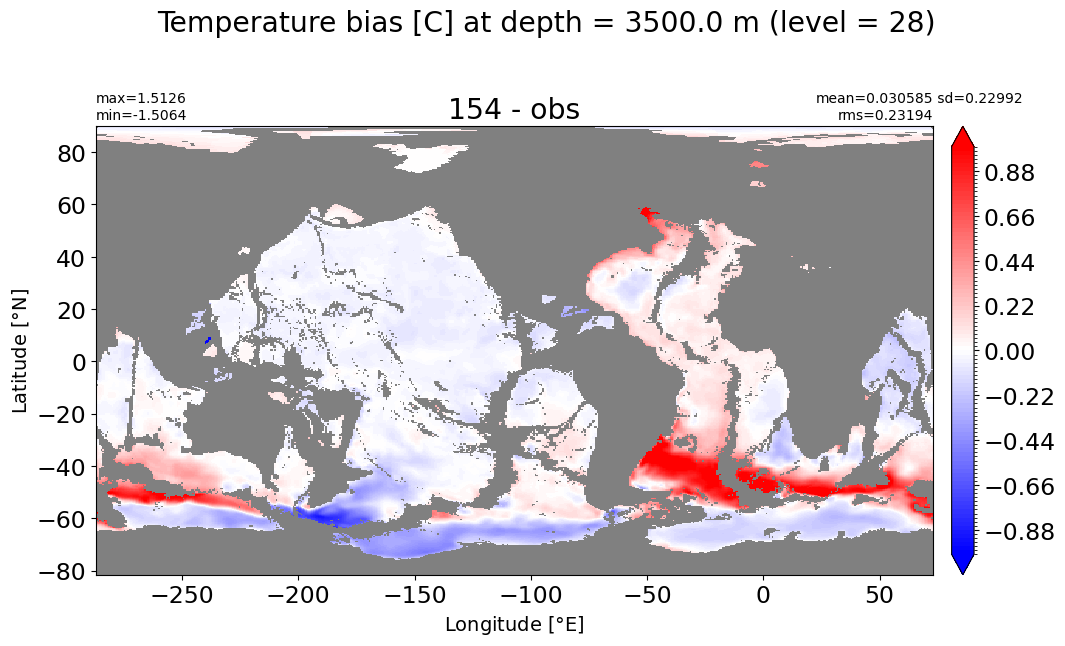
Depth = 4000 m, k=29#
make_temp_level_plot(k=29, t=1)
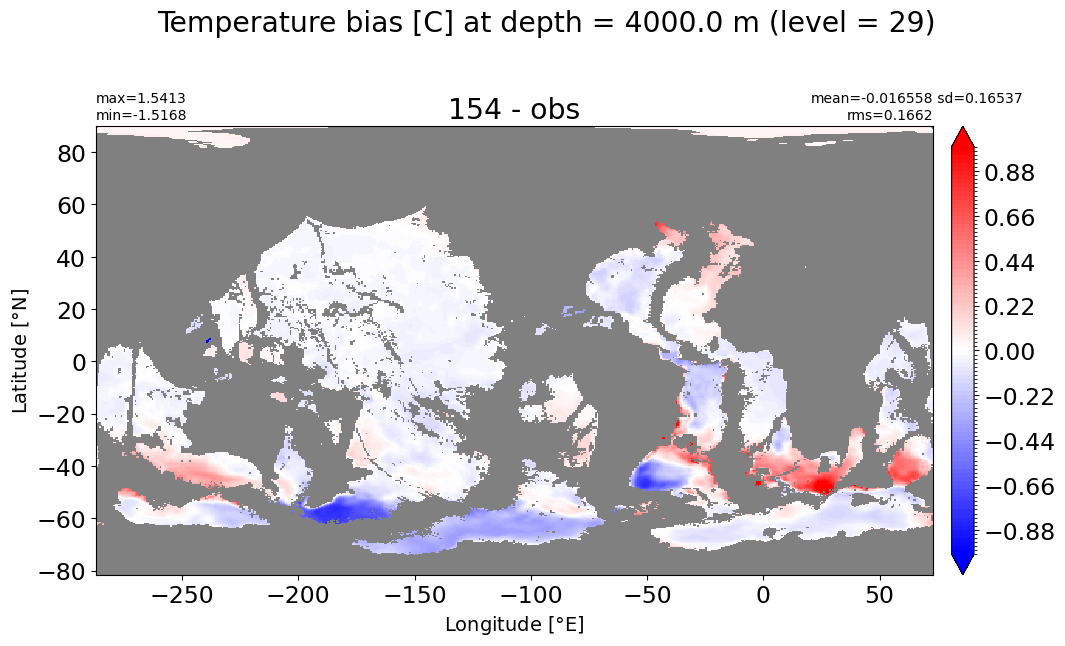
Depth = 4500 m, k=30#
make_temp_level_plot(k=30, t=1)
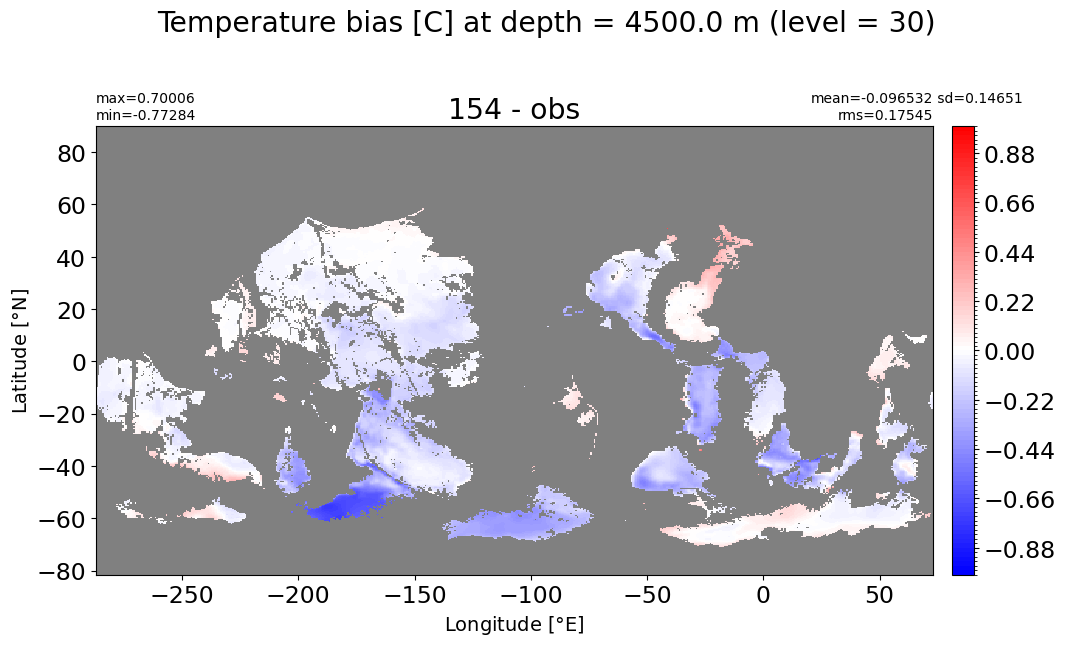
Depth = 5000 m, k=31#
make_temp_level_plot(k=31, t=0.5)
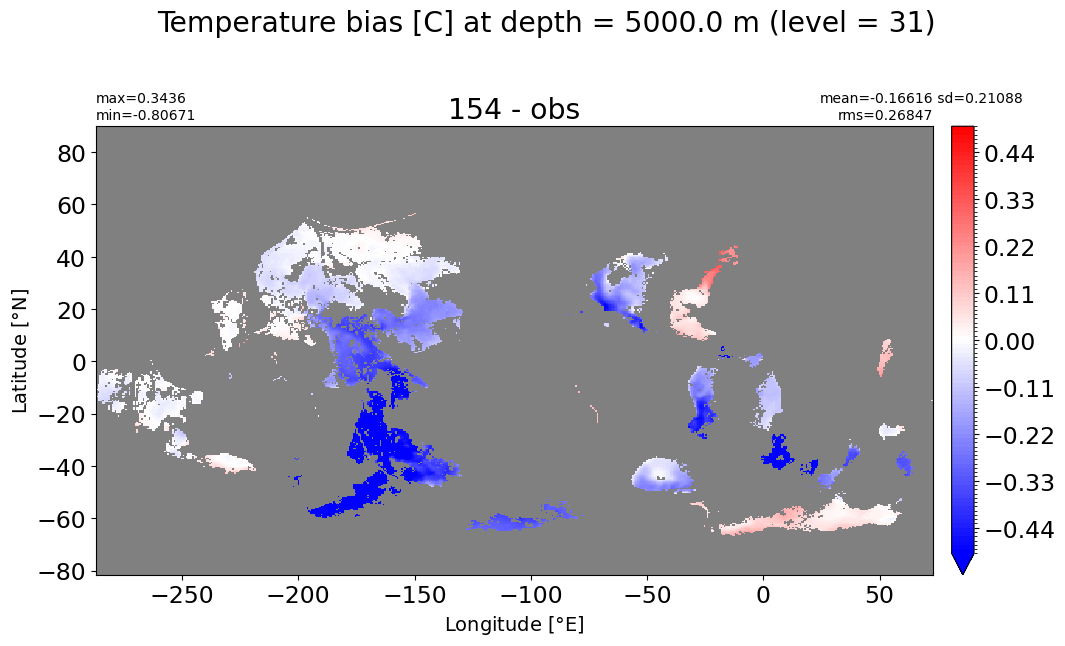
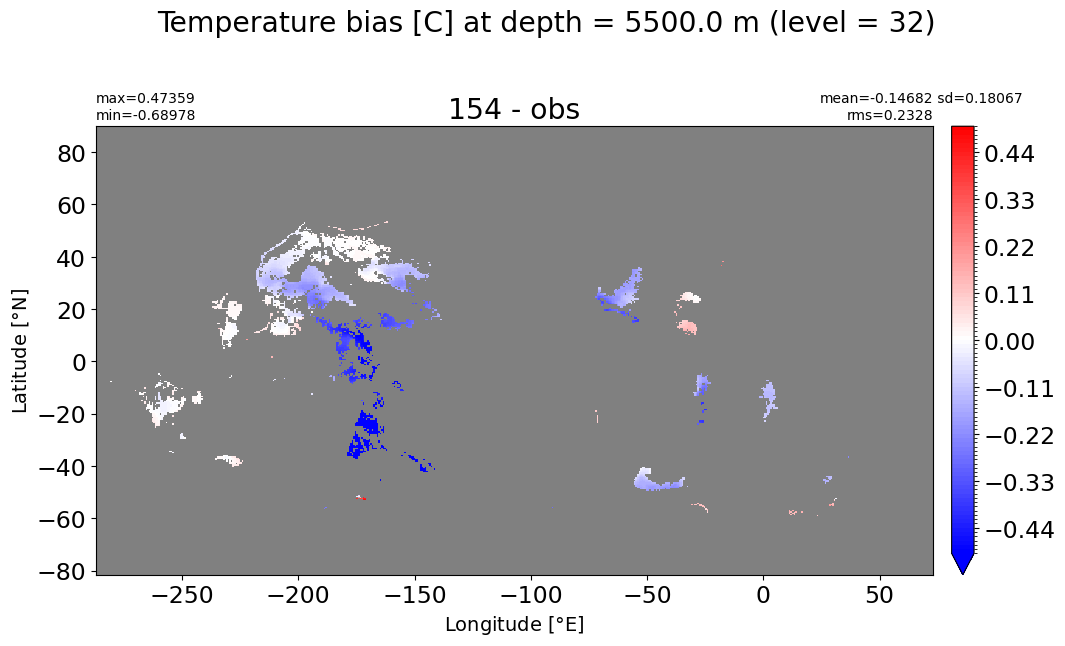
Depth = 5500 m, k=32#
make_temp_level_plot(k=32, t=0.5)
Salinity#
def make_salt_level_plot(k=0, s=5):
for path, case, i in zip(ocn_path, casename, range(len(casename))):
if 'area_t' in dir(grd[i]):
area = grd[i].area_t
else:
area = grd[i].areacello
ds_mom_s = xr.open_dataset(path+case+'_so_time_mean.nc')
fig, ax = plt.subplots(nrows=1, ncols=1, figsize=(12,6.5))
salt_mom = np.ma.masked_invalid(ds_mom_s.so_bias[k,:].values)
plt.suptitle('Salinity bias [psu] at depth = {} m (level = {})'.format(ds_mom_s.z_l[k].values, k))
xyplot(salt_mom, grd[i].geolon, grd[i].geolat, area, title=str(label[i]+' - obs'), axis=ax,
clim=(-s,s), nbins=50, colormap=plt.cm.bwr, centerlabels=True)
plt.subplots_adjust(top = 0.8)
return
Depth = 2.5 m, k=0#
make_salt_level_plot(k=0, s=5)
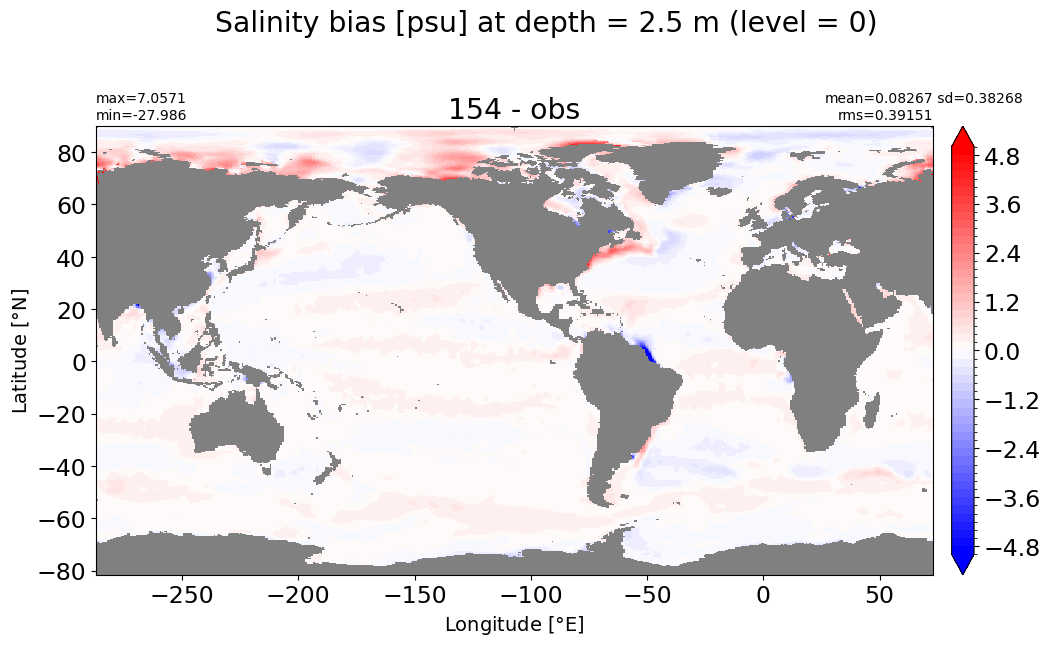
Depth = 10 m, k=1#
make_salt_level_plot(k=1, s=2.5)
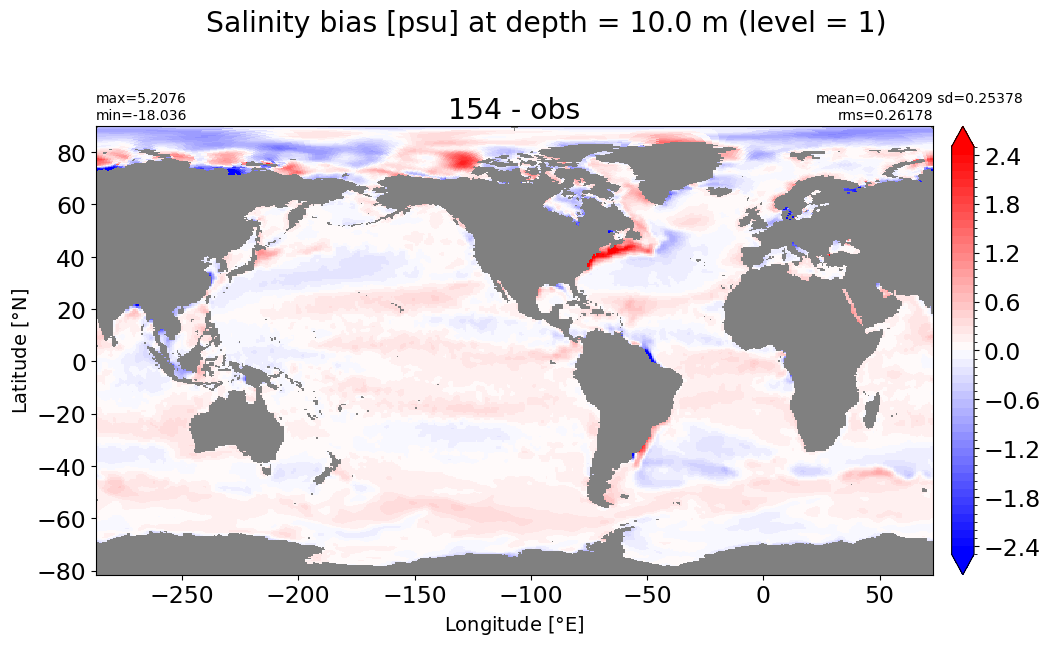
Depth = 20 m, k=2#
make_salt_level_plot(k=2, s=2.5)
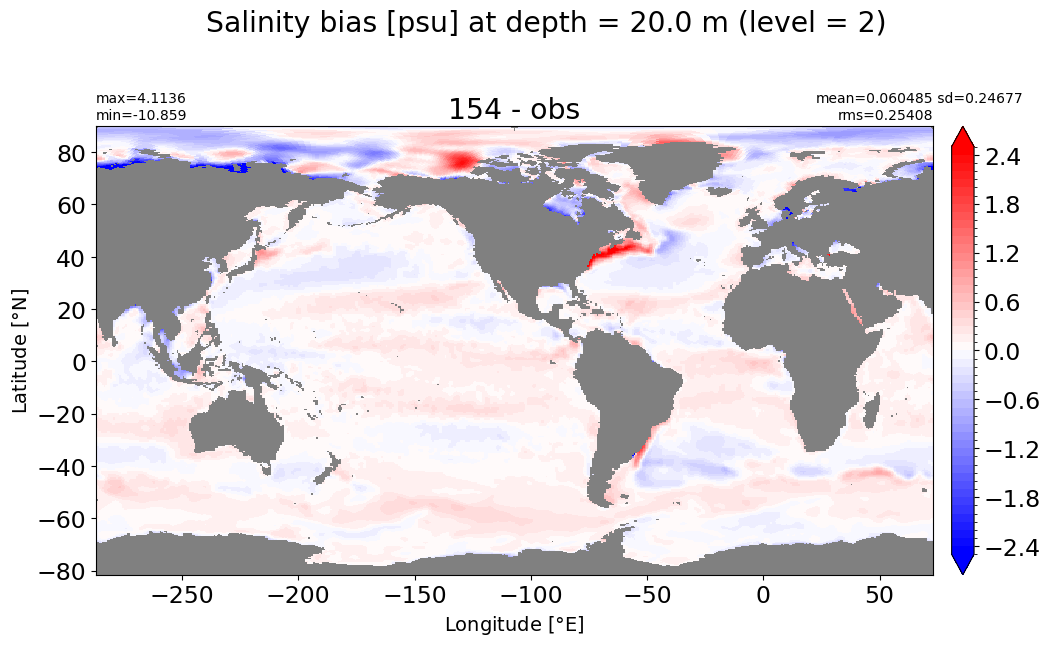
Depth = 32.5 m, k=3#
make_salt_level_plot(k=3, s=2.5)
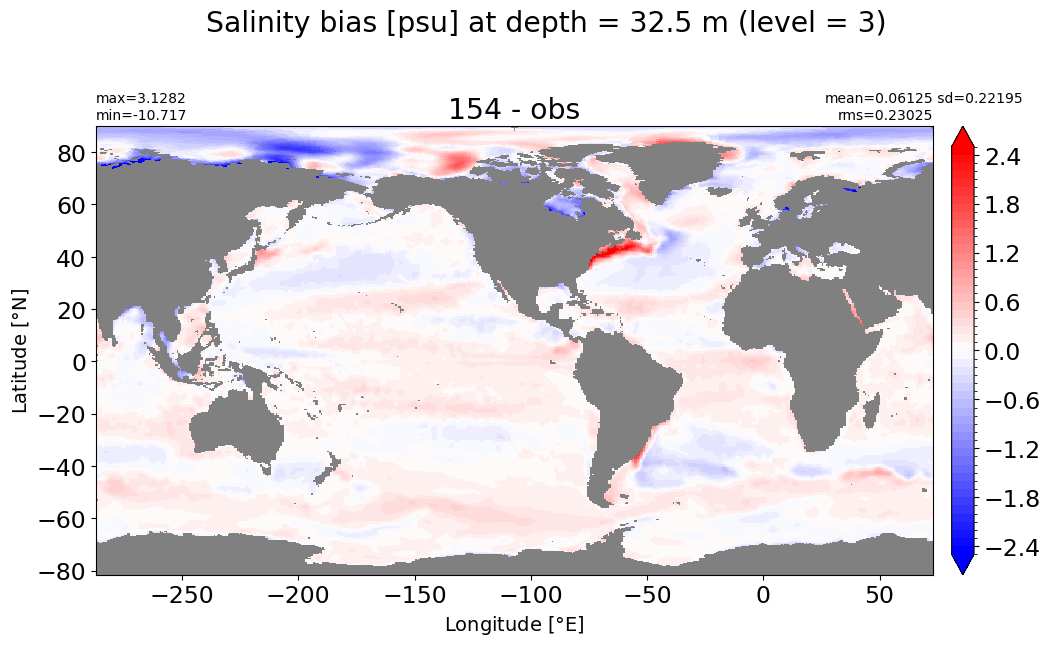
Depth = 51.25 m, k=4#
make_salt_level_plot(k=4, s=2.5)
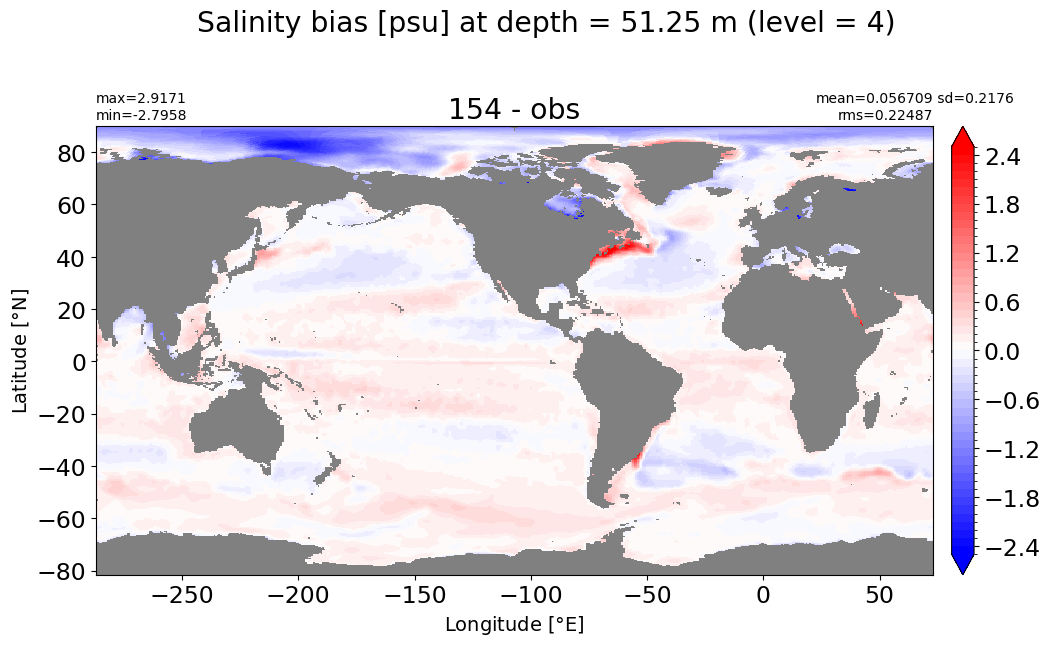
Depth = 75 m, k=5#
make_salt_level_plot(k=5, s=2.5)
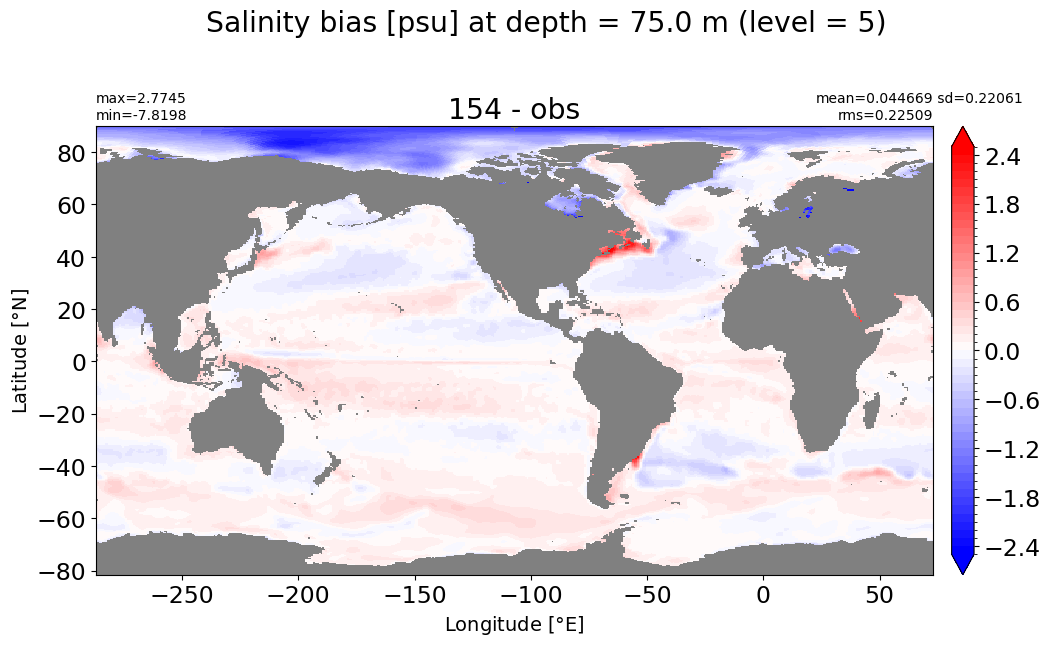
Depth = 100 m, k=6#
make_salt_level_plot(k=6, s=2.5)
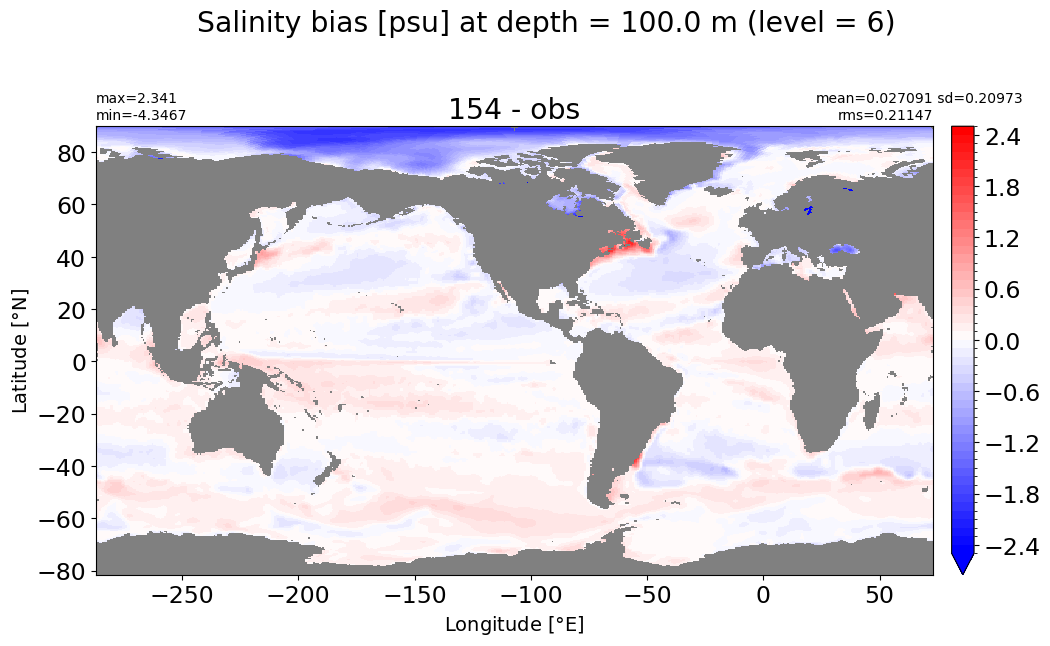
Depth = 125 m, k=7#
make_salt_level_plot(k=7, s=2.)
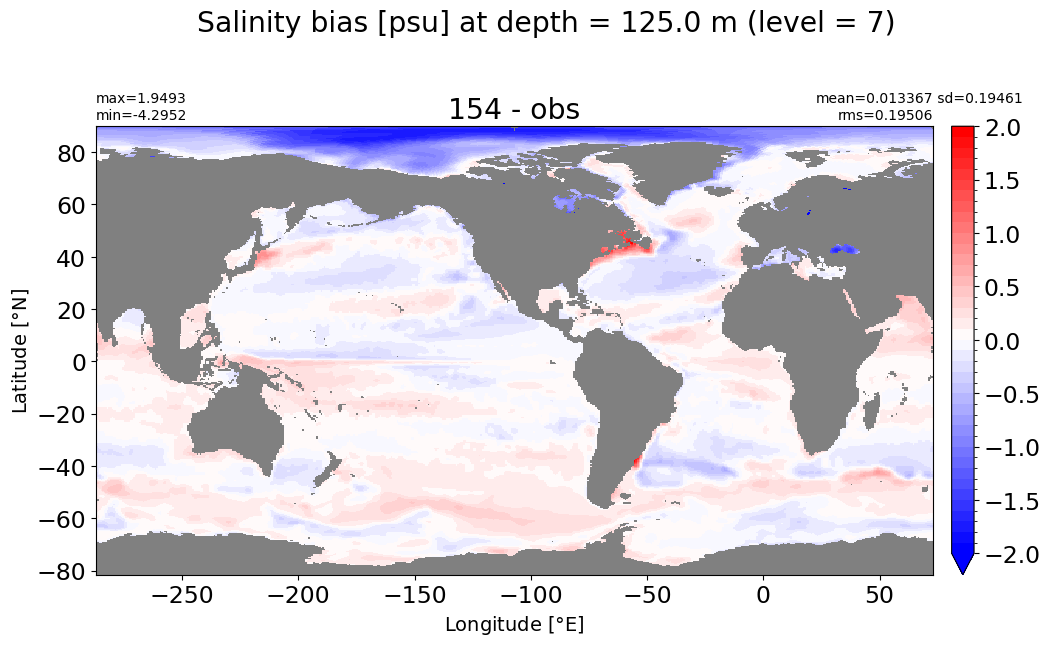
Depth = 200 m, k=9#
make_salt_level_plot(k=9, s=1.5)
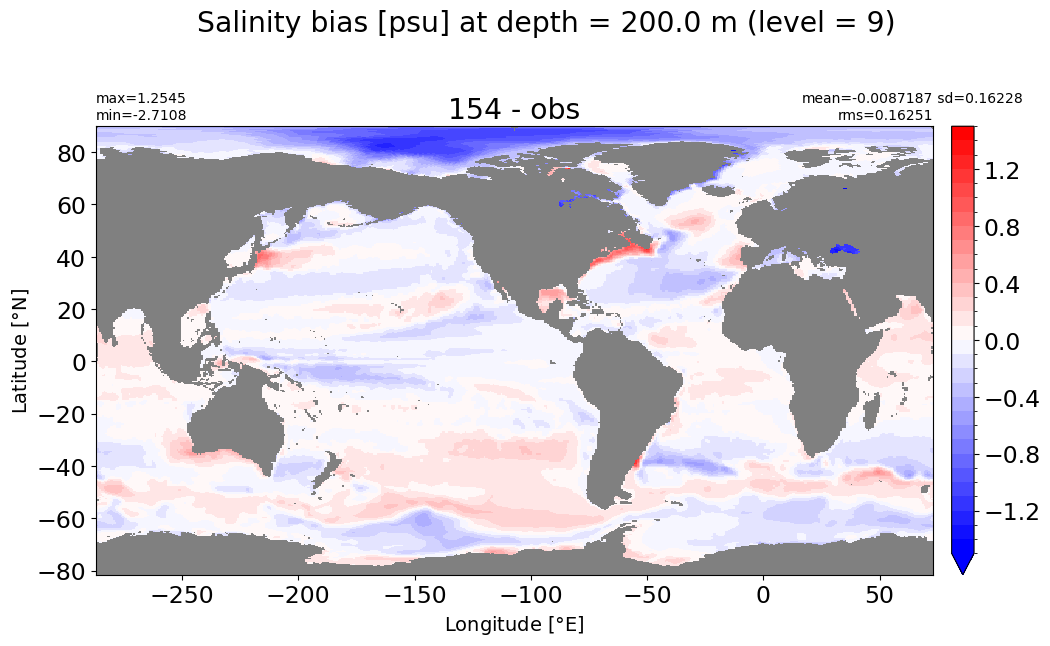
Depth = 250 m, k=10#
make_salt_level_plot(k=10, s=1.5)
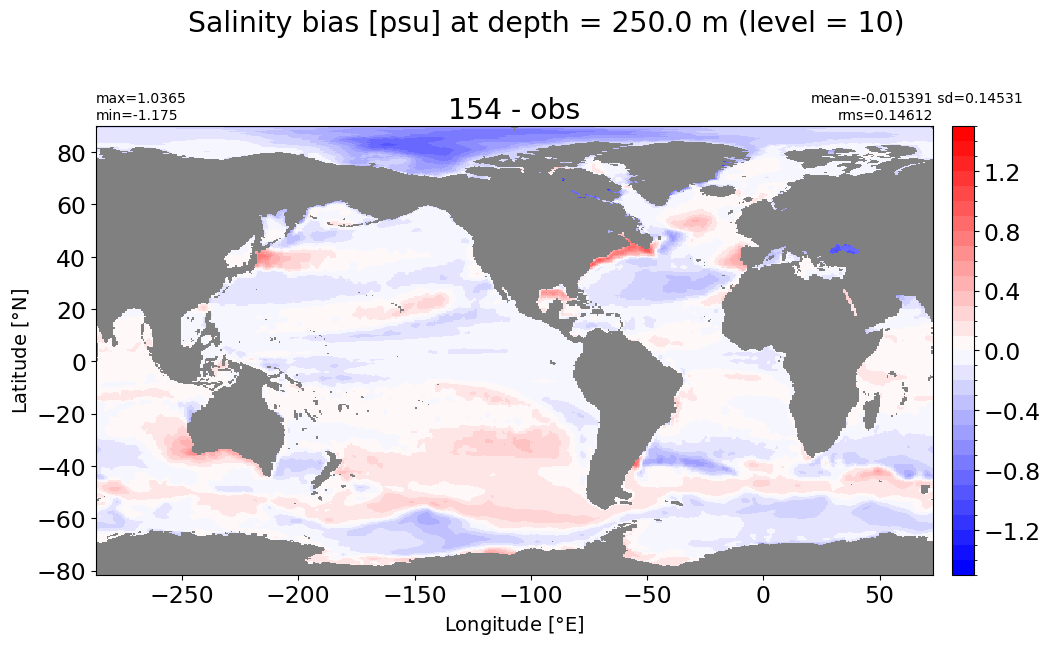
Depth = 312.25 m, k=11#
make_salt_level_plot(k=11, s=1.)
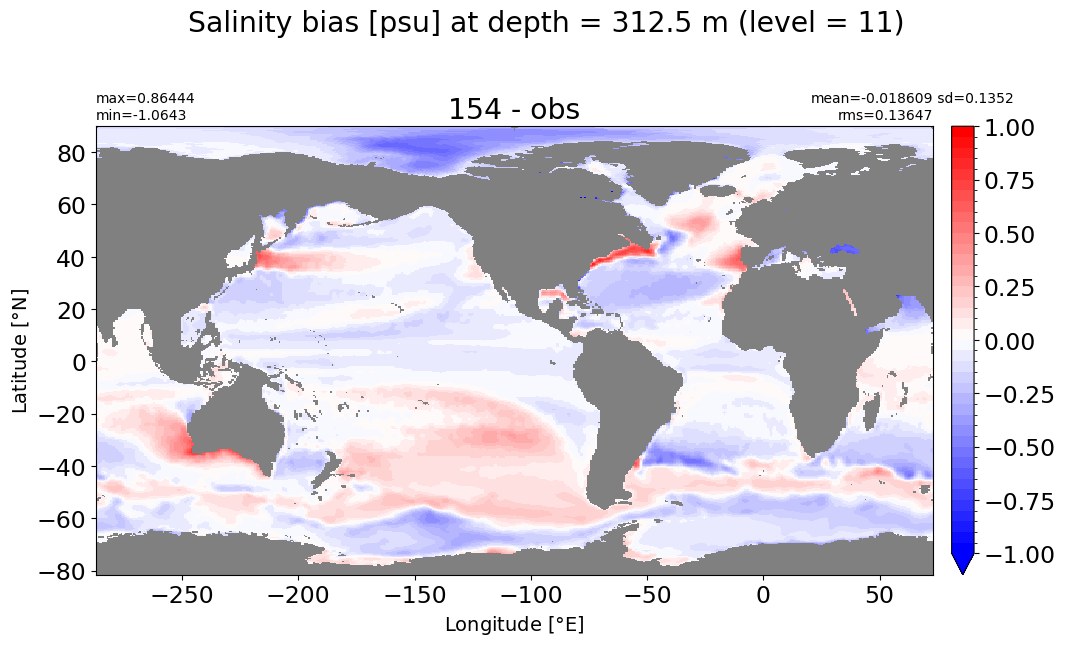
Depth = 400 m, k=12#
make_salt_level_plot(k=12, s=1.)
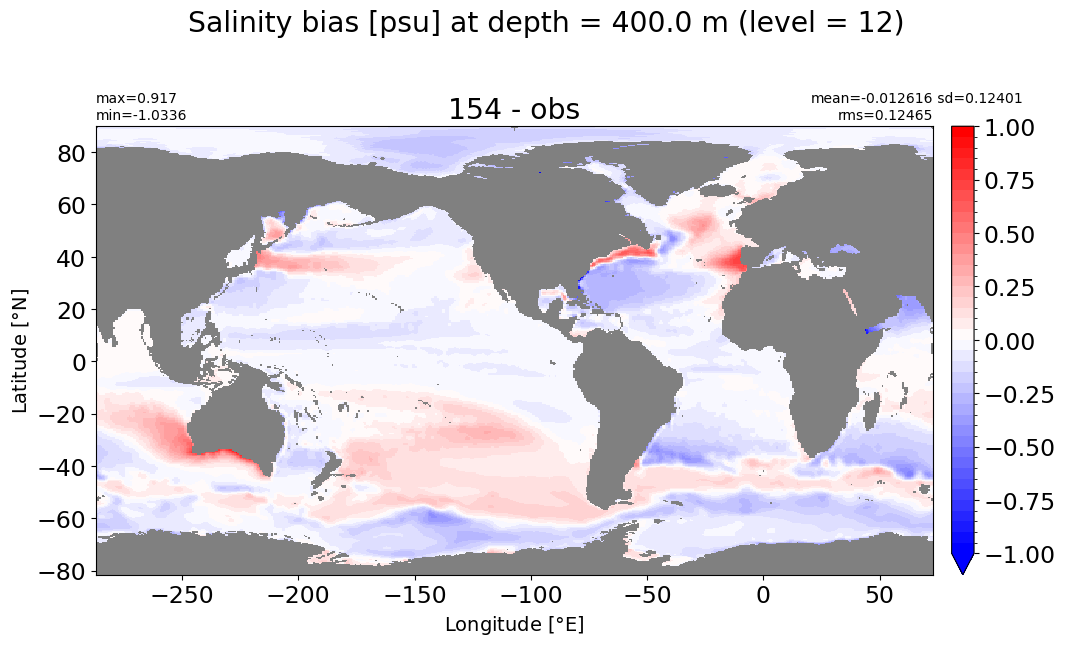
Depth = 500 m, k=13#
make_salt_level_plot(k=13, s=1.)
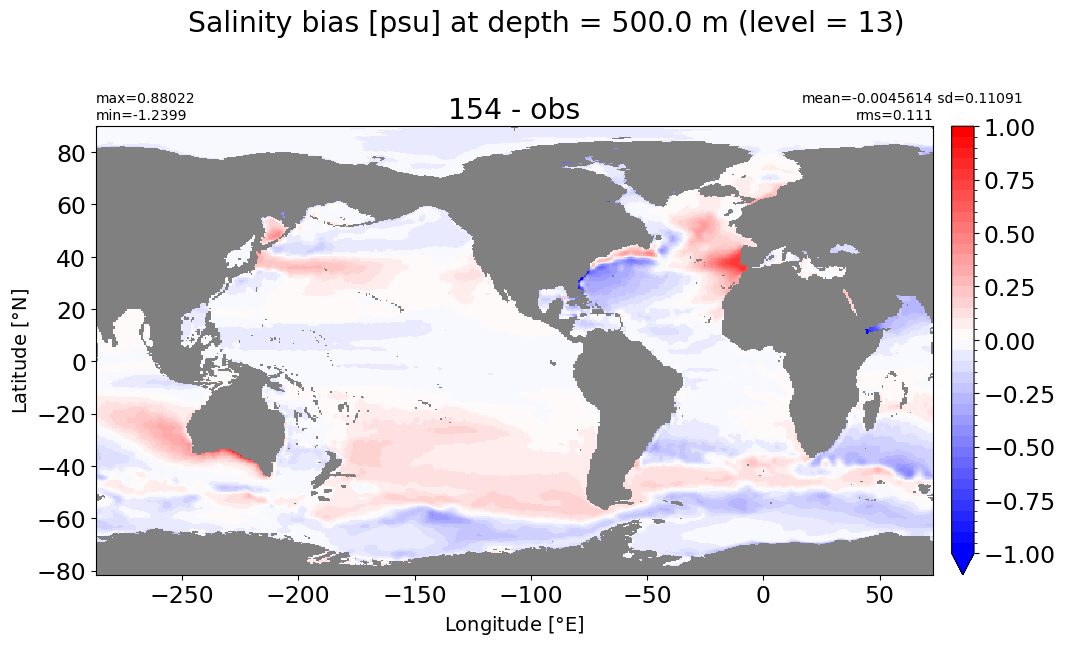
Depth = 600 m, k=14#
make_salt_level_plot(k=14, s=1.)
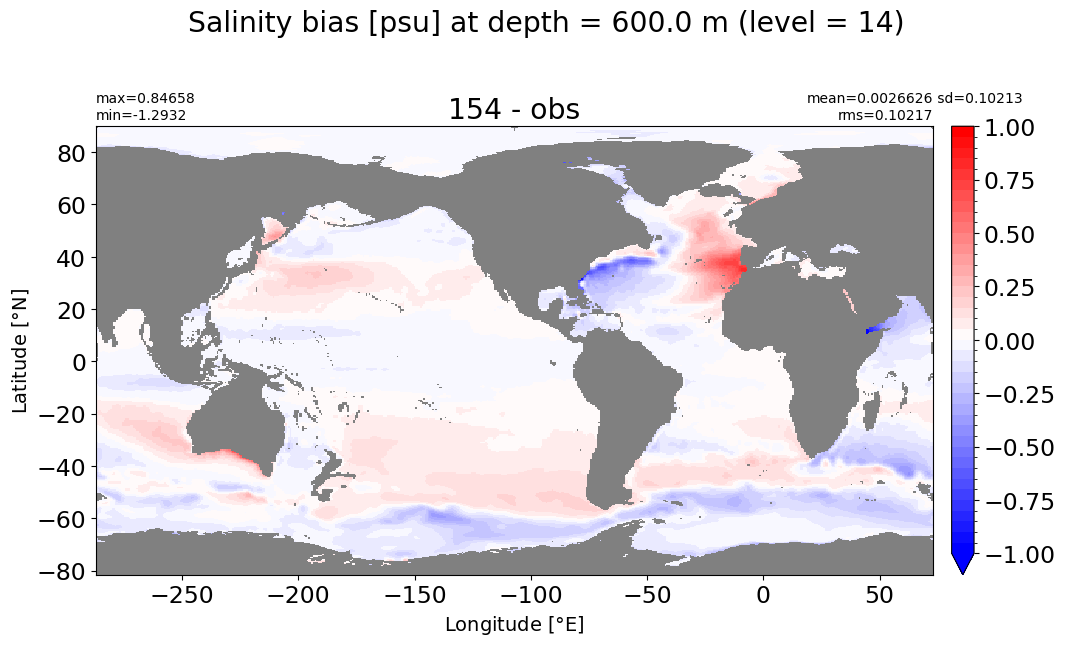
Depth = 700 m, k=15#
make_salt_level_plot(k=15, s=1.)
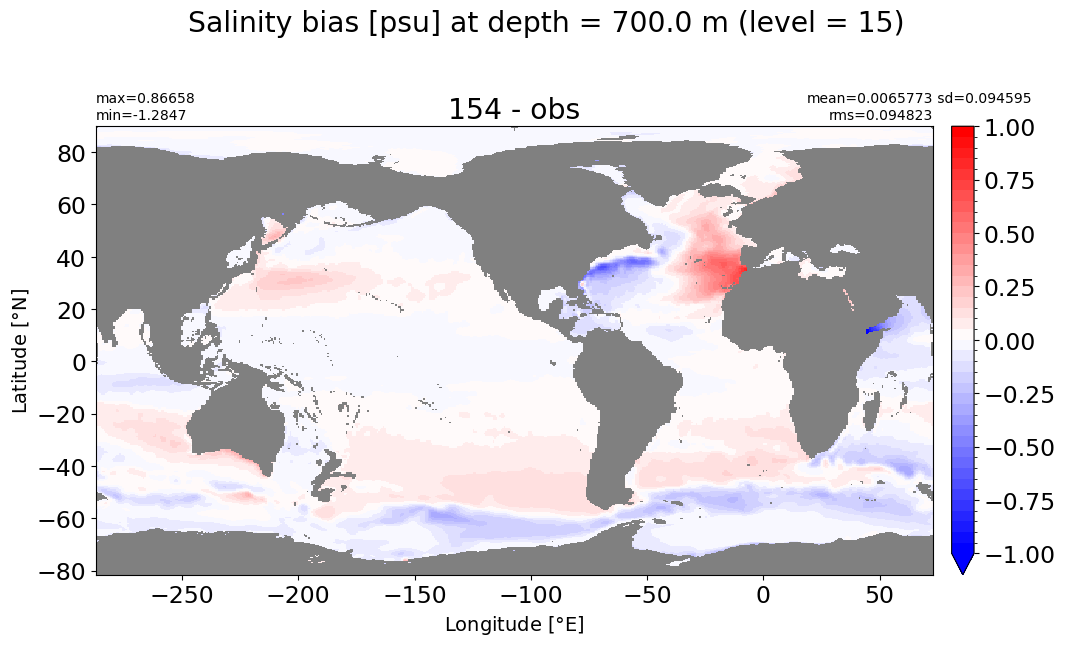
Depth = 800 m, k=16#
make_salt_level_plot(k=16, s=0.5)
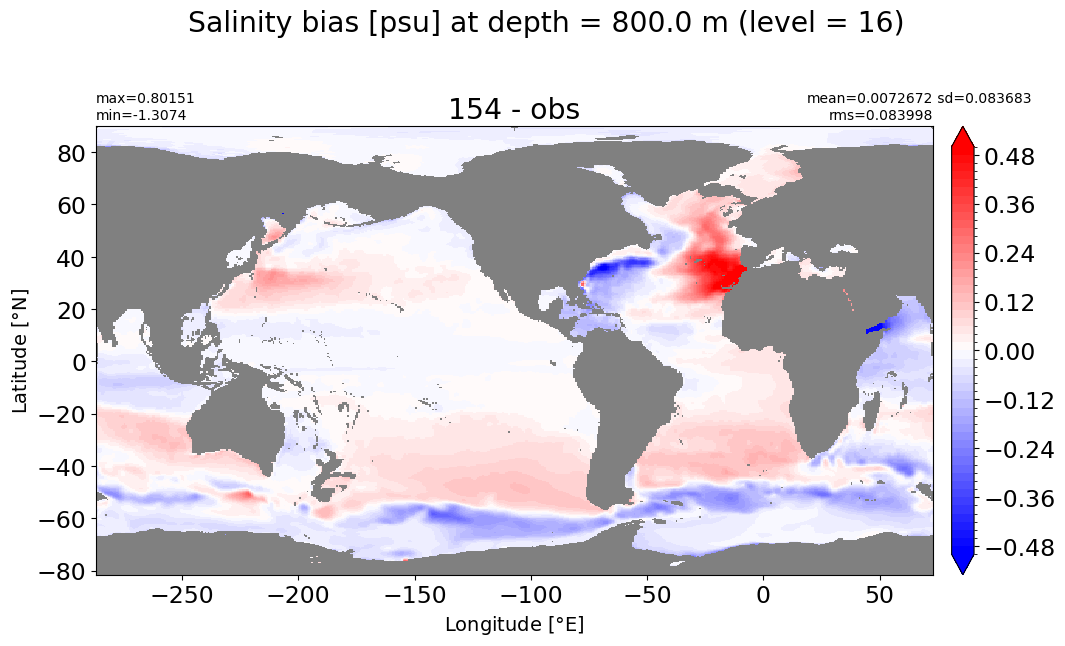
Depth = 900 m, k=17#
make_salt_level_plot(k=17, s=0.5)
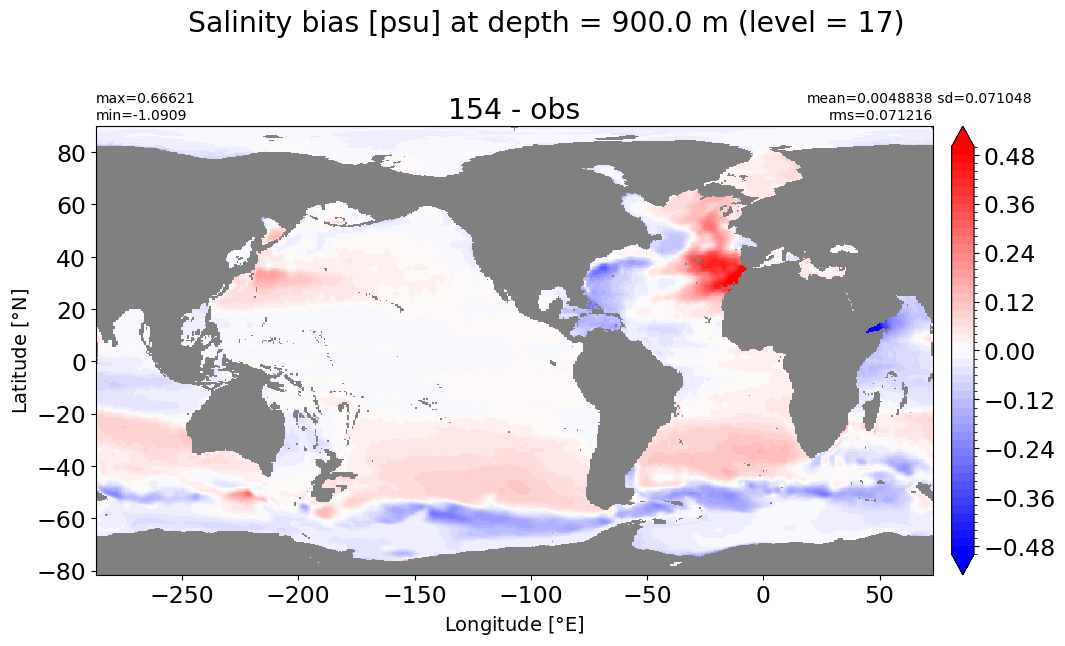
Depth = 1000 m, k=18#
make_salt_level_plot(k=18, s=0.5)
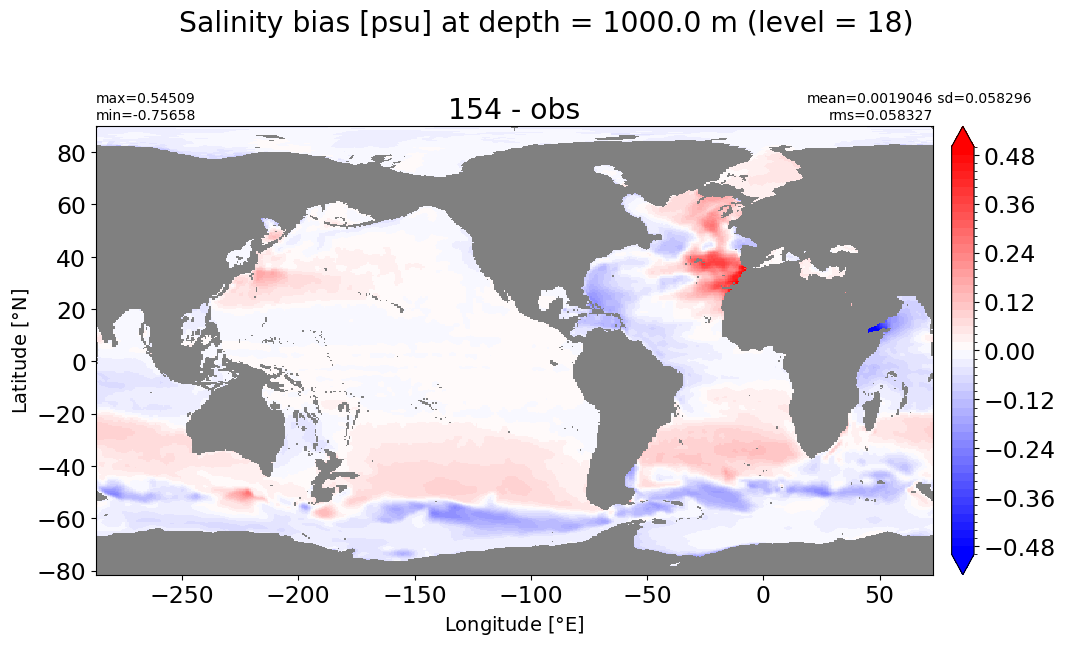
Depth = 1100 m, k=19#
make_salt_level_plot(k=19, s=0.5)
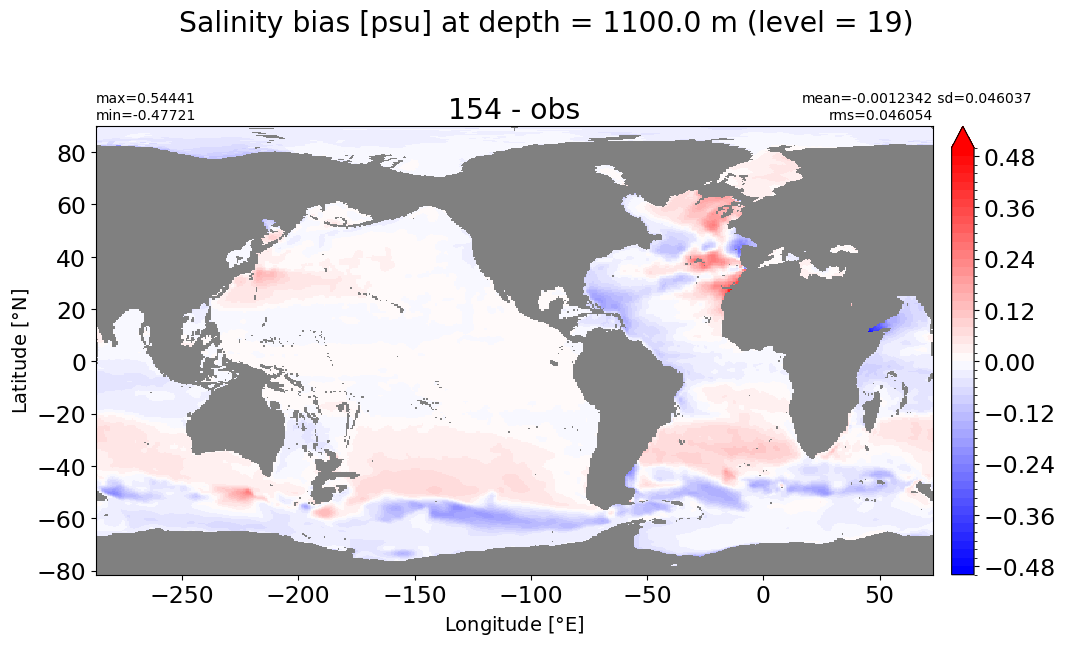
Depth = 1200 m, k=20#
make_salt_level_plot(k=20, s=0.25)
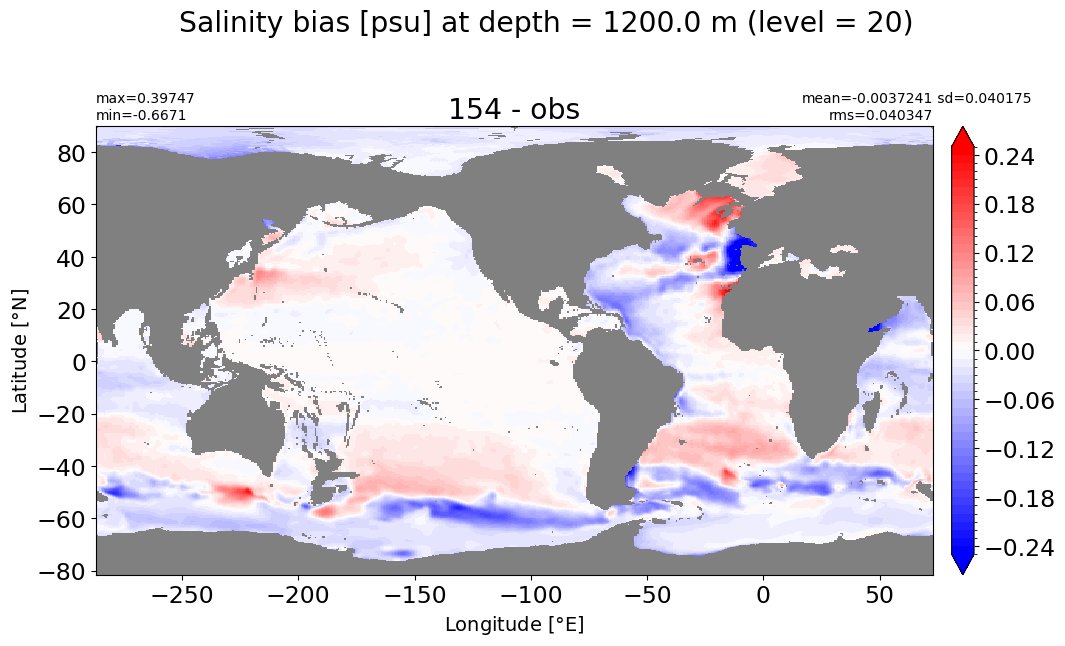
Depth = 1300 m, k=21#
make_salt_level_plot(k=21, s=0.25)
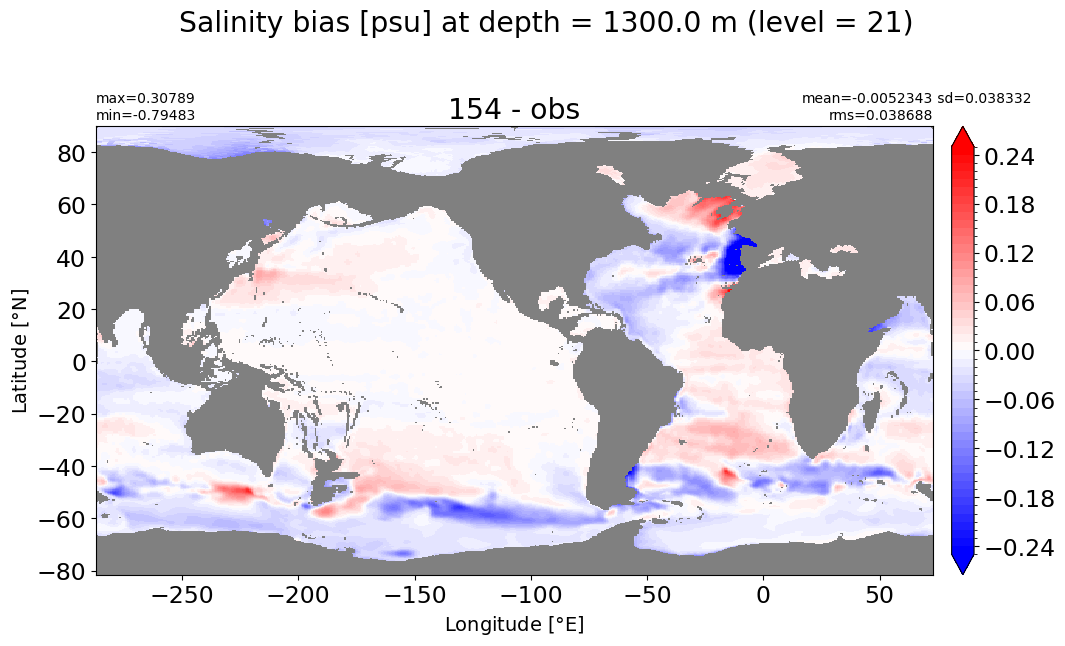
Depth = 1400 m, k=22#
make_salt_level_plot(k=22, s=0.25)
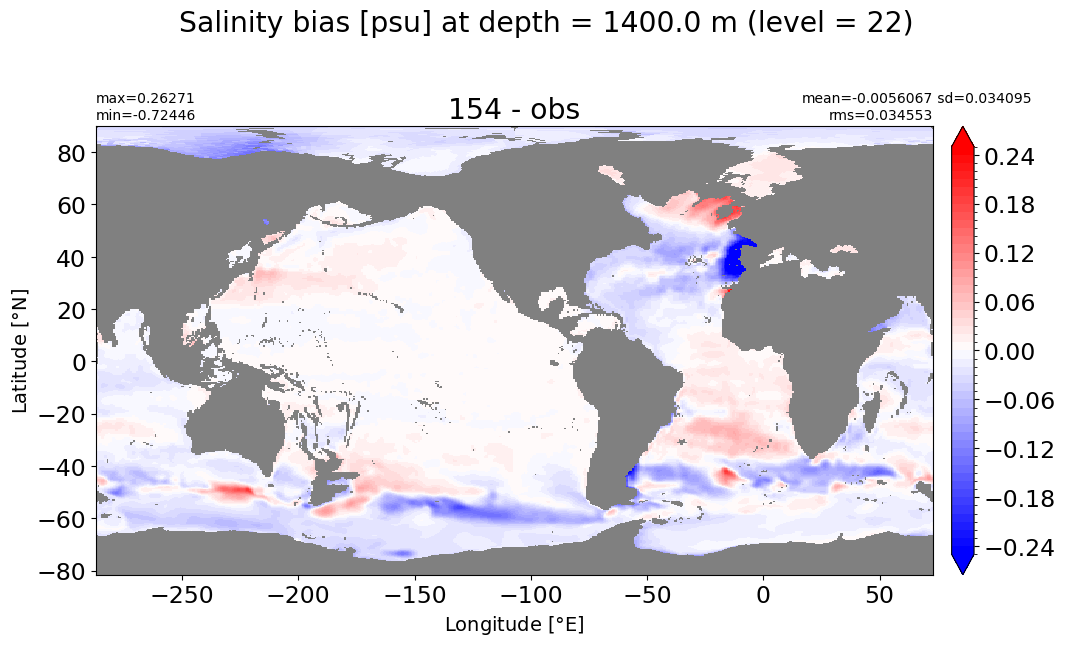
Depth = 1537.5 m, k=23#
make_salt_level_plot(k=23, s=0.2)
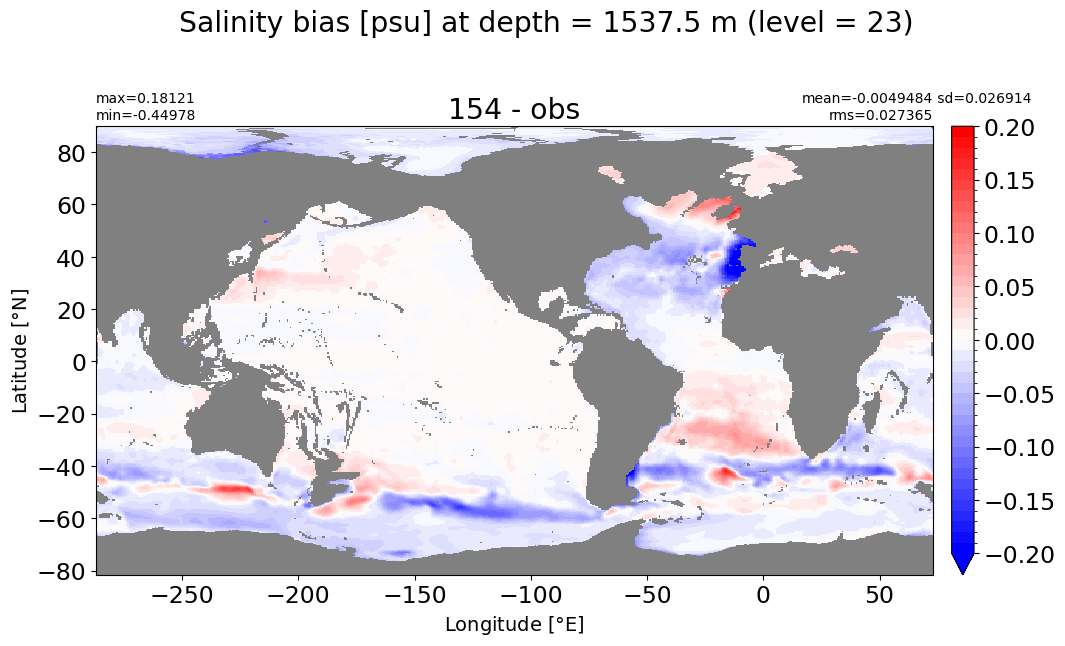
Depth = 1750 m, k=24#
make_salt_level_plot(k=24, s=0.2)
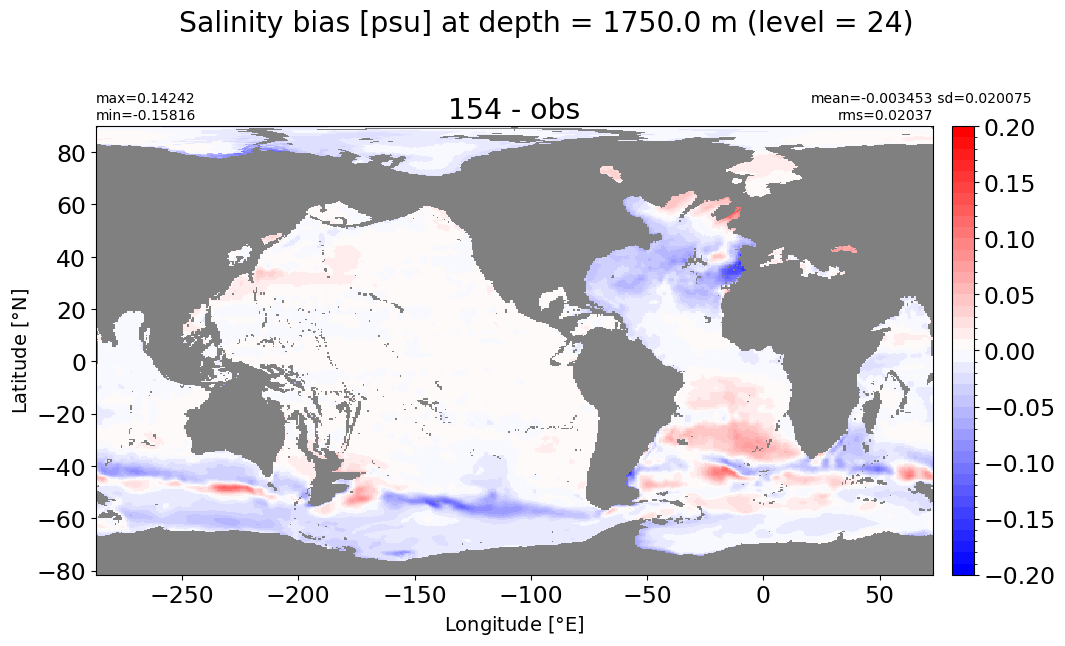
Depth = 2062.5 m, k=25#
make_salt_level_plot(k=25, s=0.1)
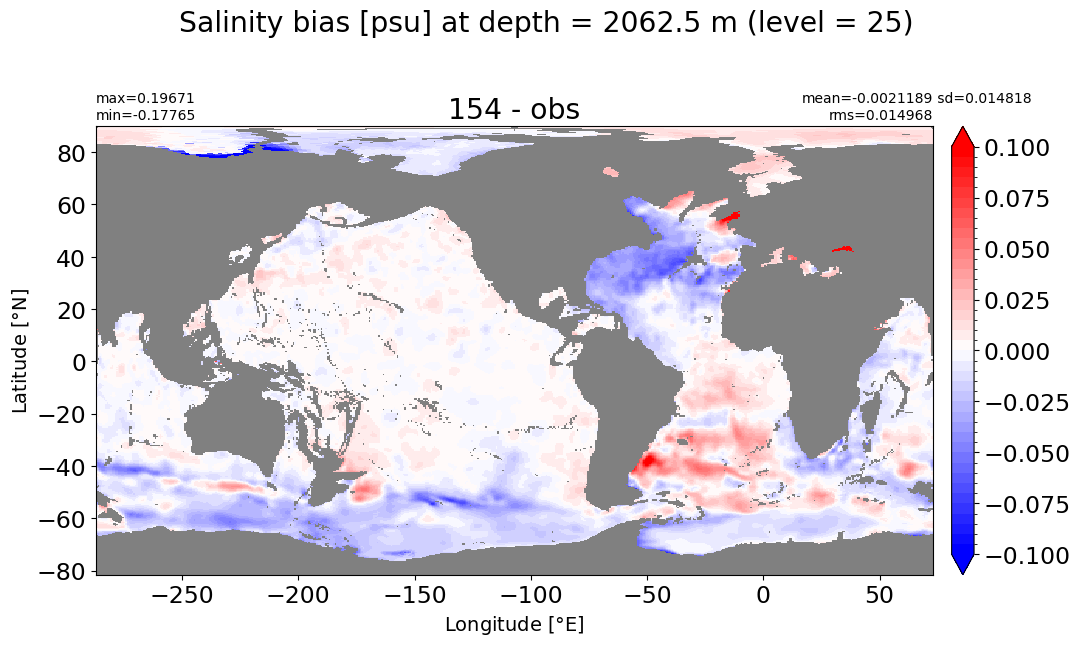
Depth = 2500 m, k=26#
make_salt_level_plot(k=26, s=0.1)
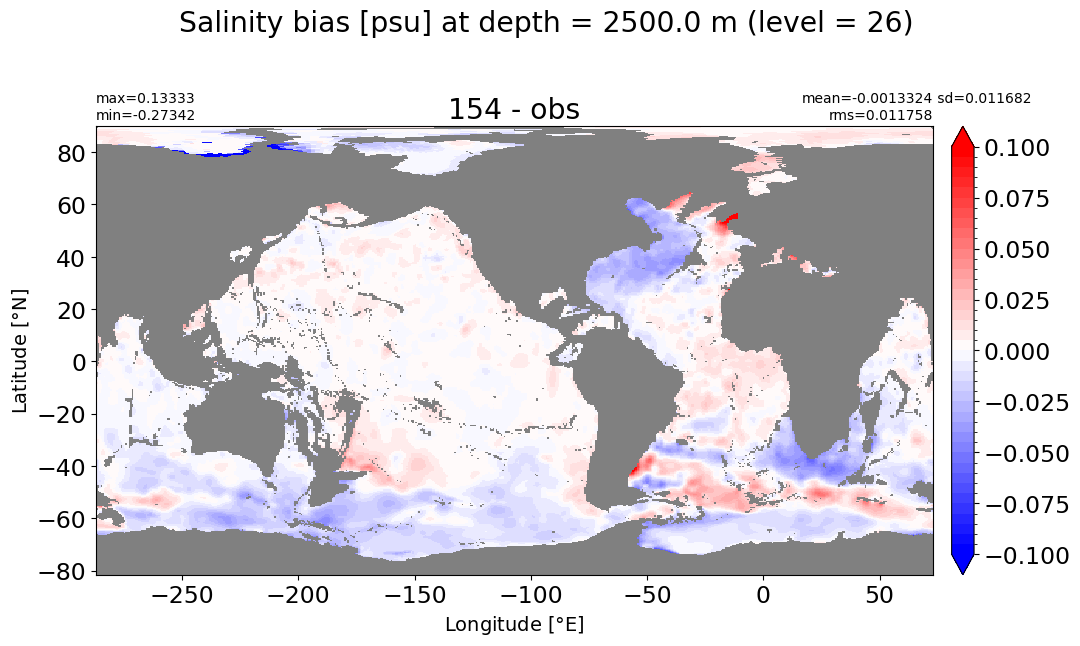
Depth = 3000 m, k=27#
make_salt_level_plot(k=27, s=0.1)
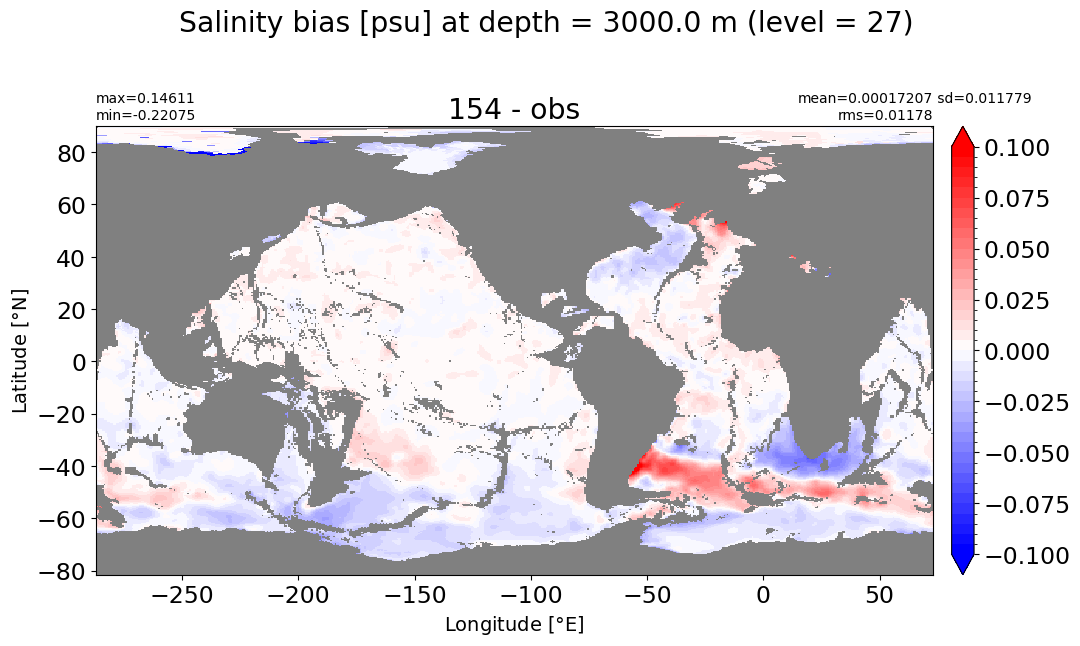
Depth = 3500 m, k=28#
make_salt_level_plot(k=28, s=0.1)
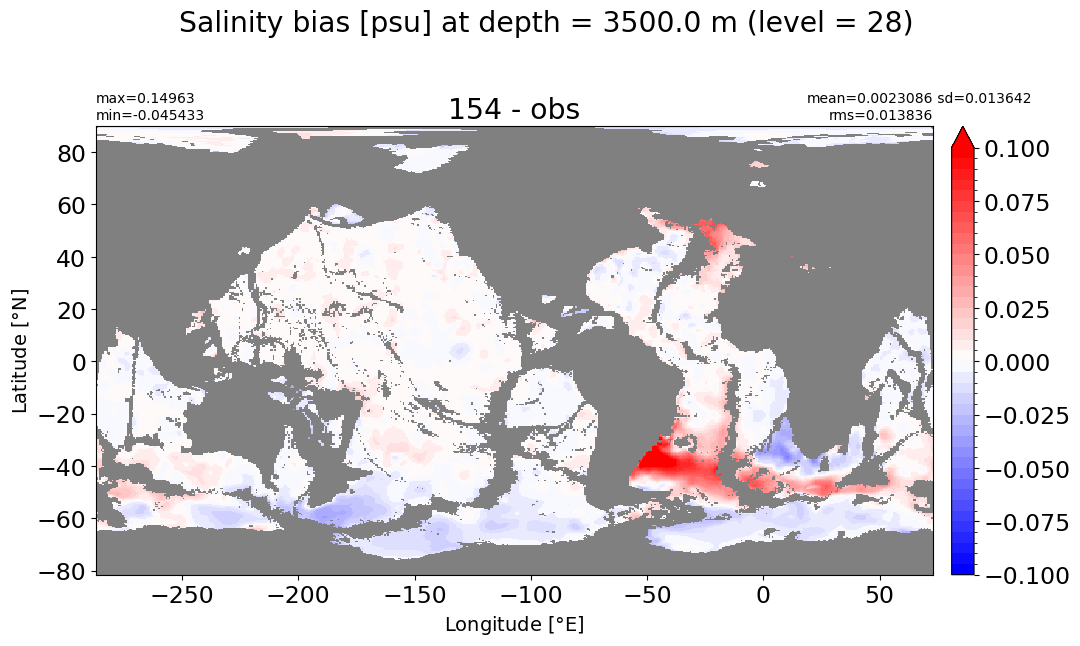
Depth = 4000 m, k=29#
make_salt_level_plot(k=29, s=0.1)
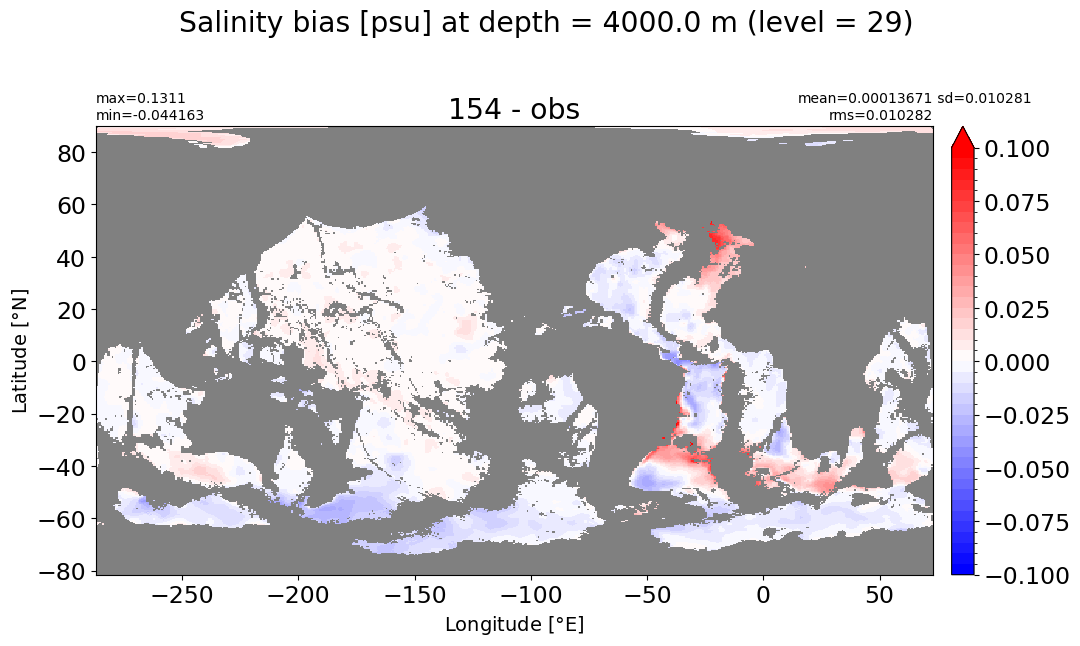
Depth = 4500 m, k=30#
make_salt_level_plot(k=30, s=0.05)
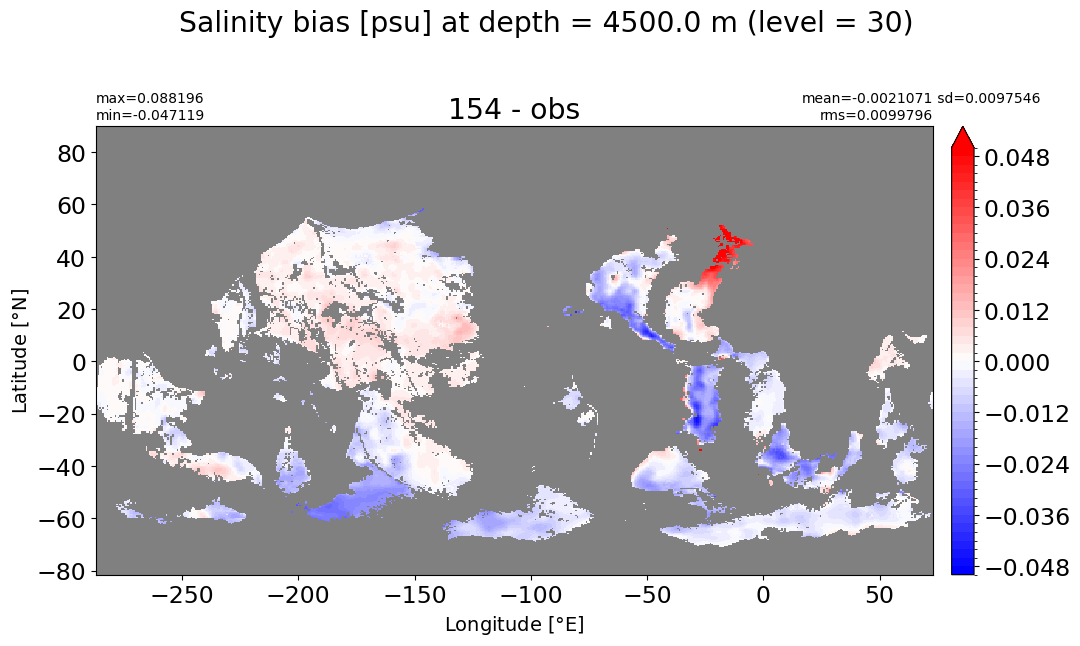
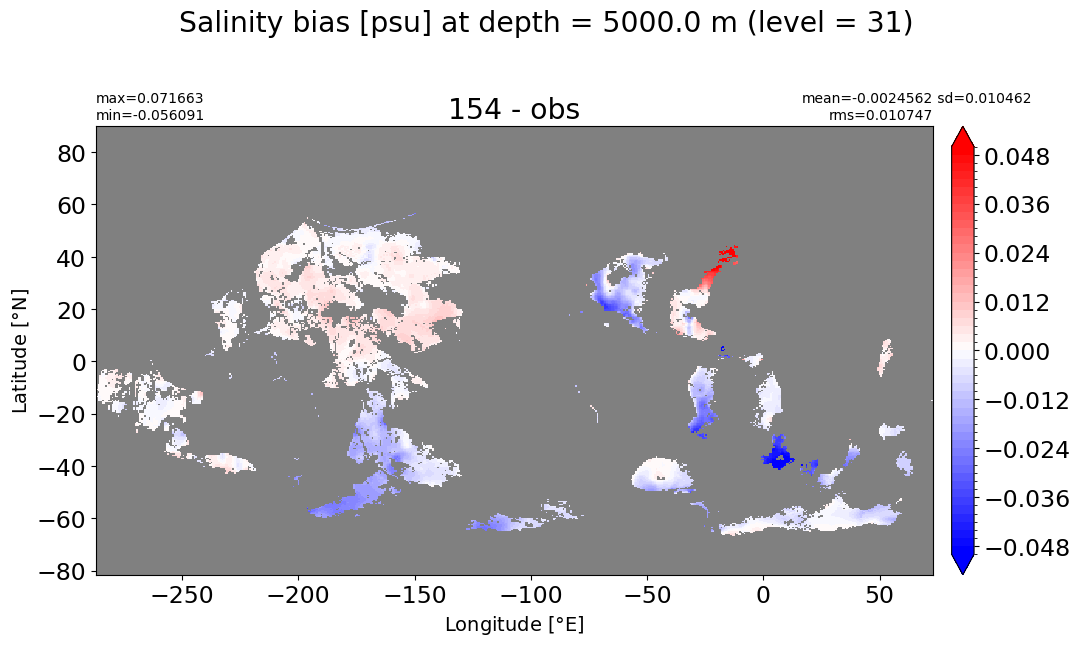
Depth = 5000 m, k=31#
make_salt_level_plot(k=31, s=0.05)
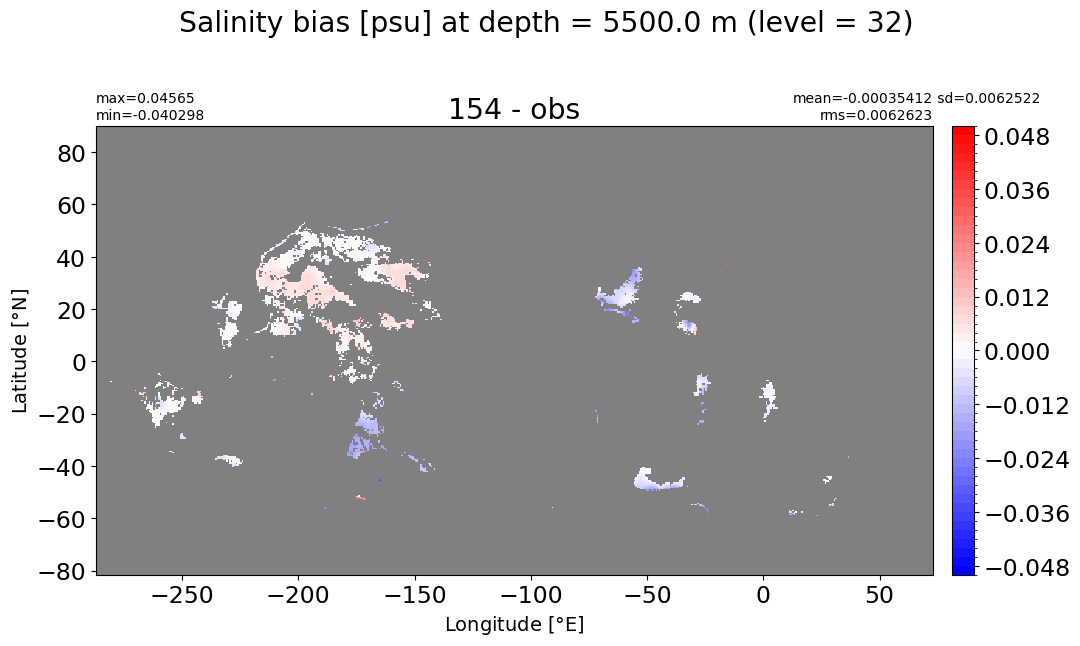
Depth = 5500 m, k=32#
make_salt_level_plot(k=32, s=0.05)
Zonal biases#
reg_mom = ['Global', 'PersianGulf', 'RedSea', 'BlackSea', 'MedSea', 'BalticSea',
'HudsonBay', 'Arctic', 'PacificOcean', 'AtlanticOcean', 'IndianOcean',
'SouthernOcean', 'LabSea', 'BaffinBay', 'EGreenlandIceland',
'GulfOfMexico', 'Maritime', 'SouthernOcean60S']
Potential temperature#
reg = "AtlanticOcean"
var = 'thetao_bias'
units = r'$^o$C'
regions = ['Global','MedSea','Arctic','PacificOcean',
'AtlanticOcean','IndianOcean','SouthernOcean',
'LabSea']
ylim_min = [-90, 30, 62, -50, -50, -50, -90, 50]
ylim_max = [ 90, 47, 90, 65, 72, 30, -35, 68 ]
cmap = plt.get_cmap('bwr')
cmap.set_bad(color='black')
matplotlib.rcParams.update({'font.size': 16})
for reg, j in zip(regions, range(len(regions))):
da = []
for path, case, i in zip(ocn_path, casename, range(len(casename))):
ds_mom_t = xr.open_dataset(path+case+'_thetao_time_mean.nc')
atl = basin_code[i].sel(region=reg)
# Set zero values to NaN
atl = atl.where(atl != 0, np.nan)
da.append((ds_mom_t[var] * atl).weighted(grd_xr[i].areacello.fillna(0)).mean('xh').expand_dims(case=[label[i]]).sel(yh=slice(ylim_min[j],
ylim_max[j])))
combined_da = xr.concat(da, dim="case")
col = None
if len(combined_da) > 1:
col = "case"
g = combined_da.plot(x="yh", y="z_l", col=col,
yincrease=False, col_wrap=2,
figsize=(12,4), cmap=cmap, robust=True,
cbar_kwargs={"label": reg + ', ' + var + ' ({})'.format(units)})
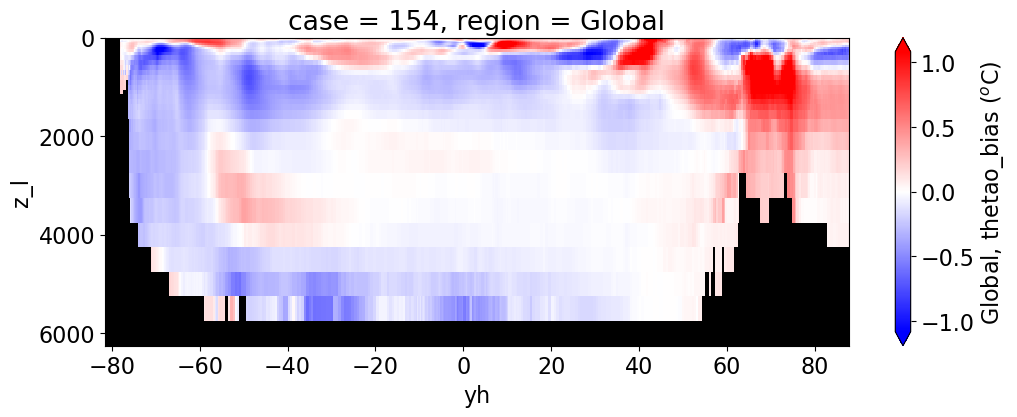
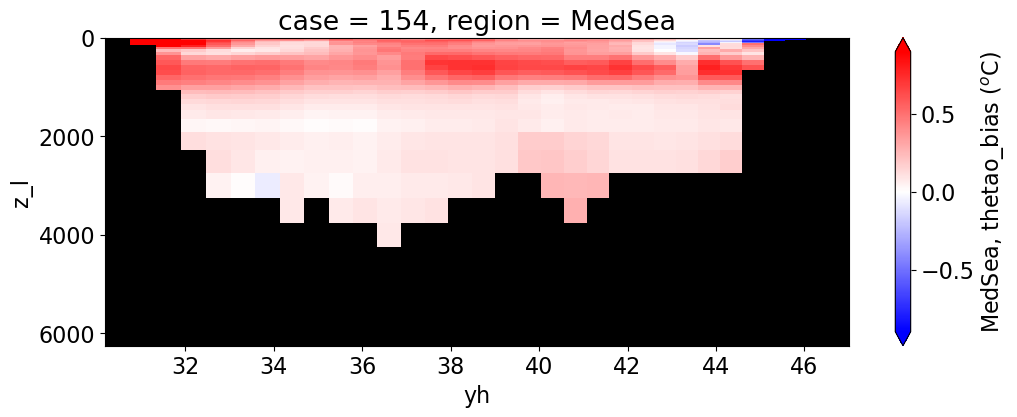
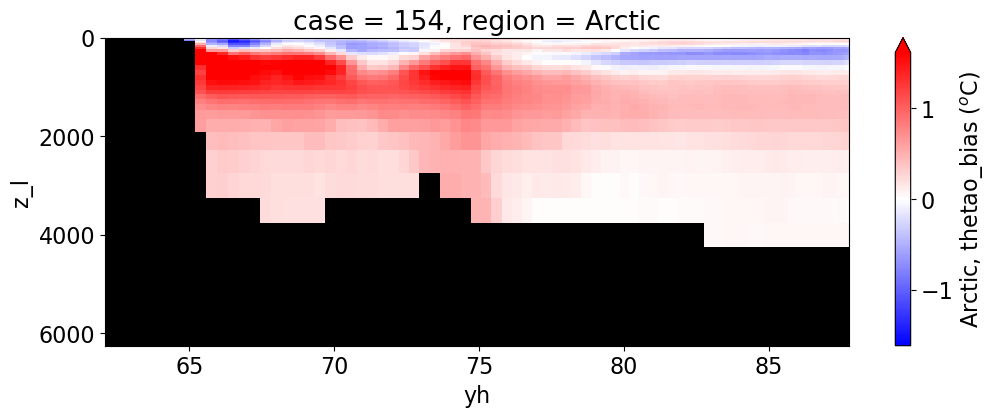
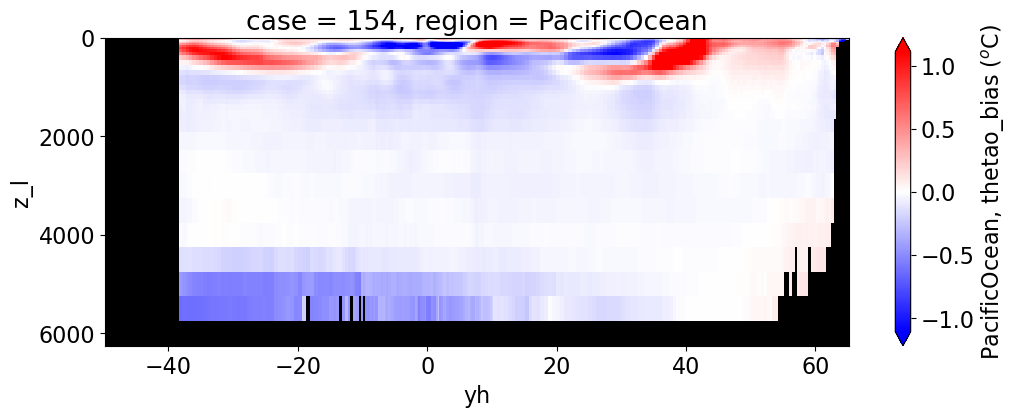
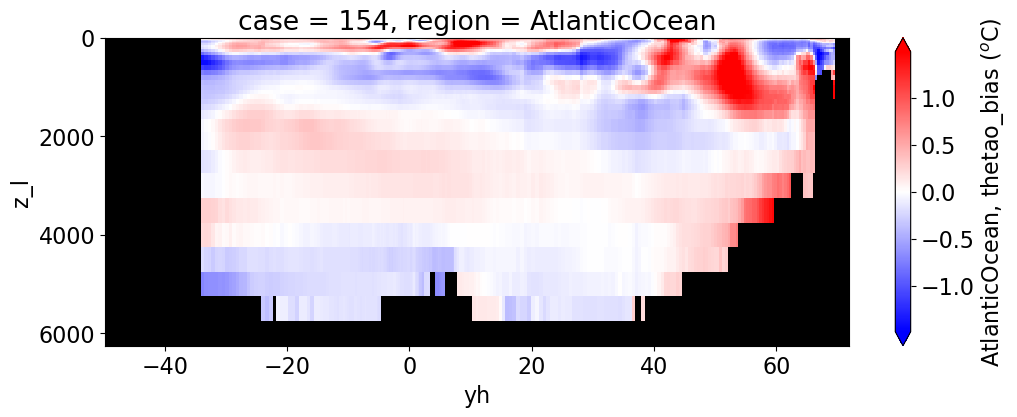
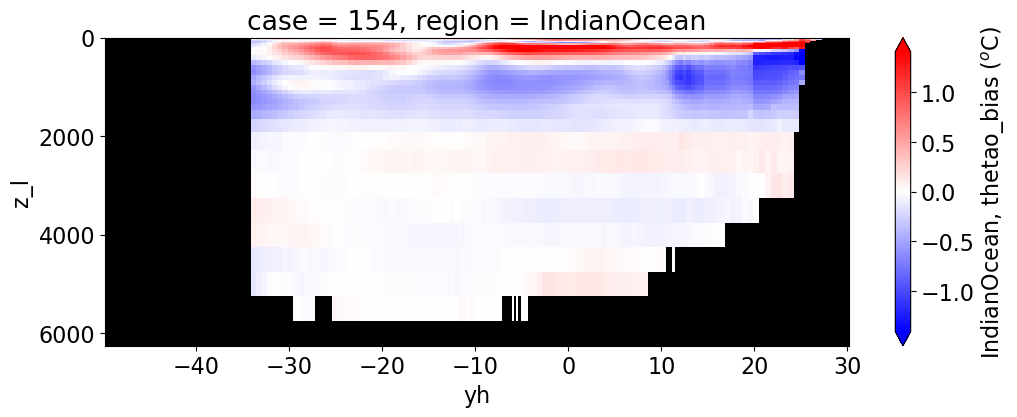
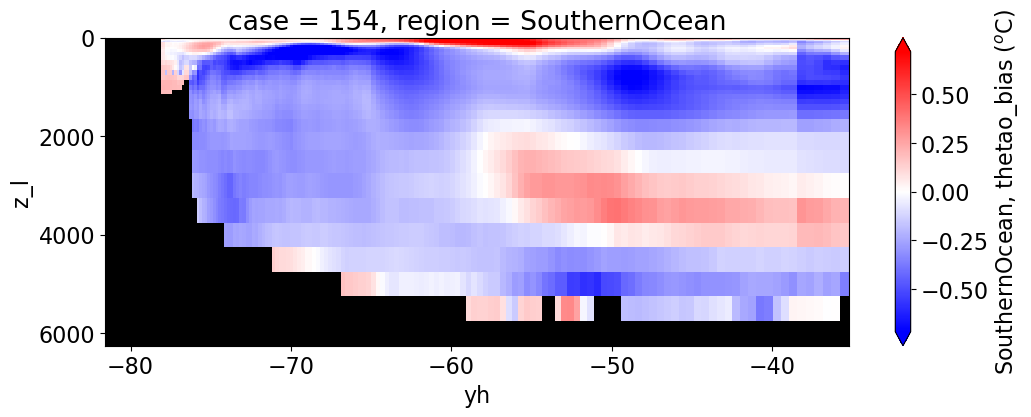
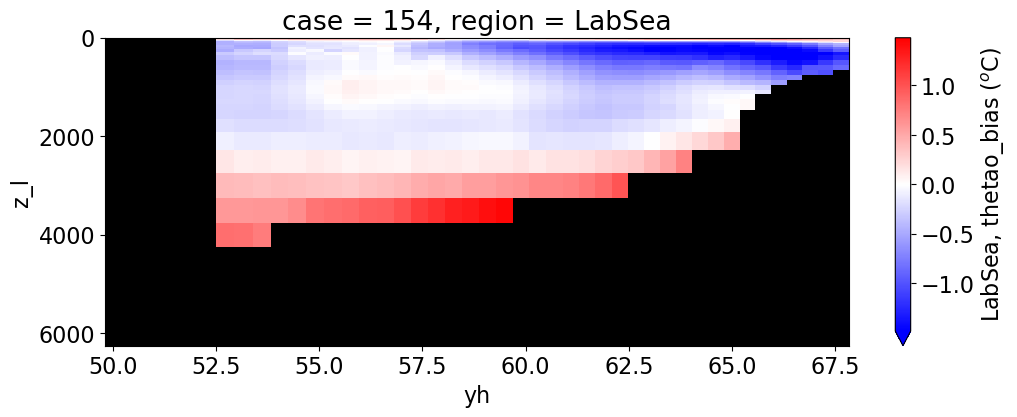
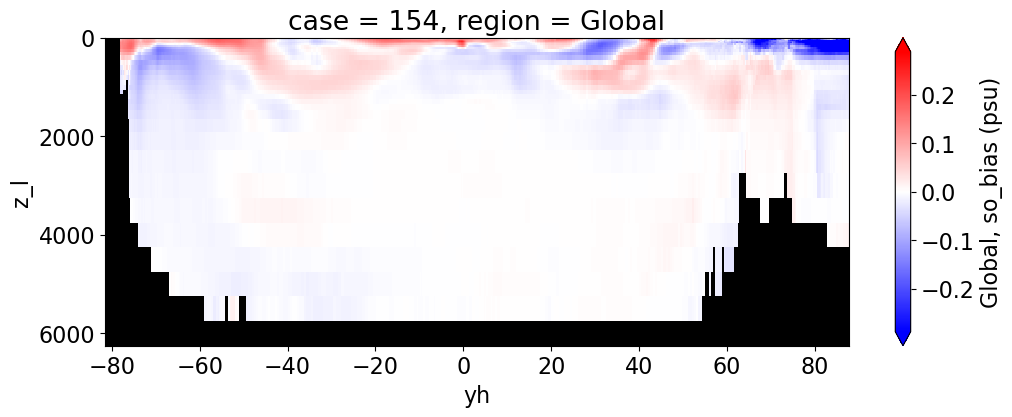
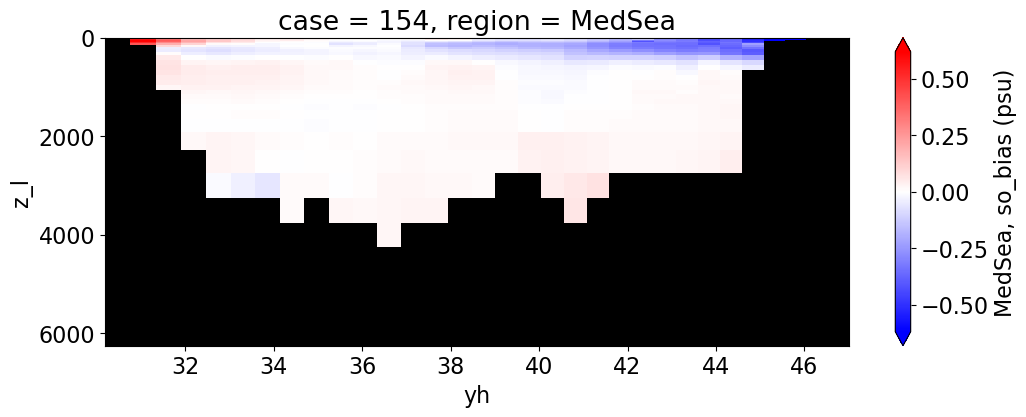
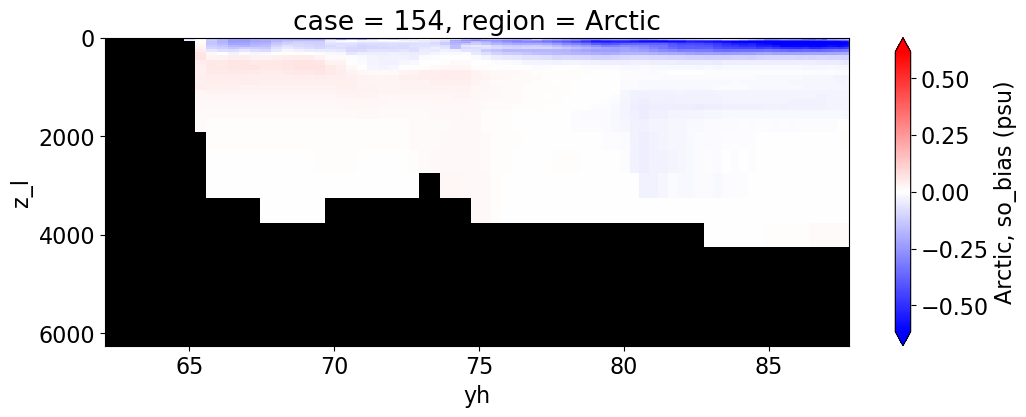
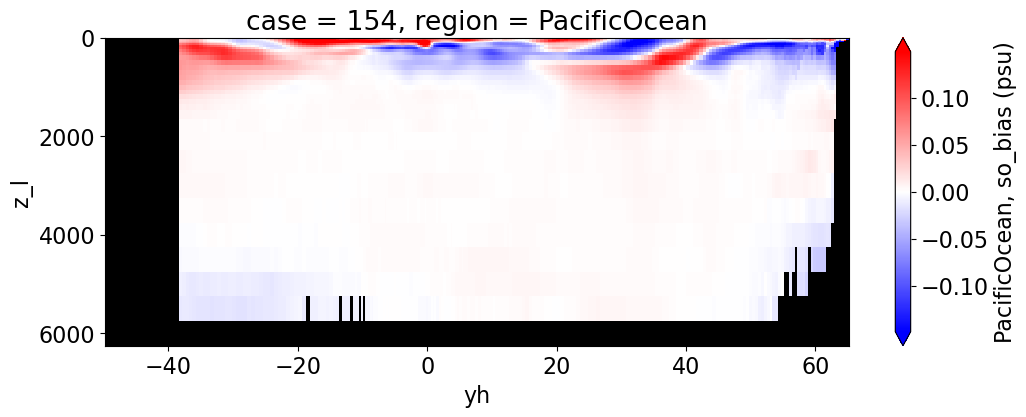
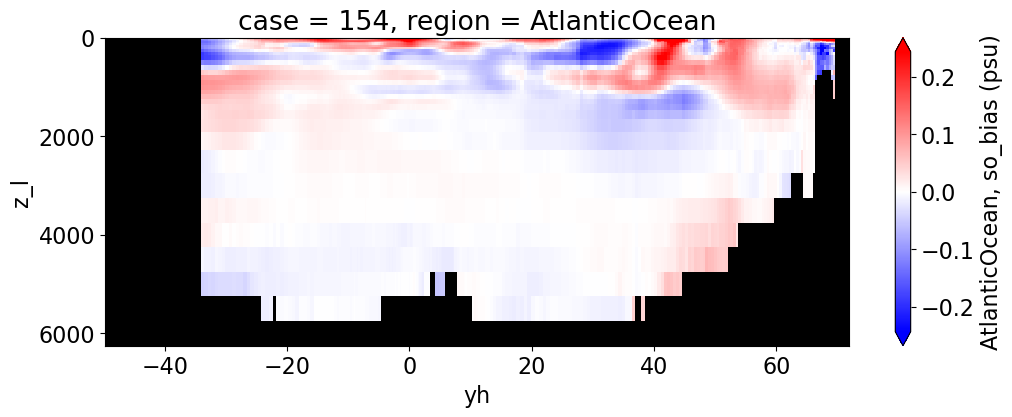
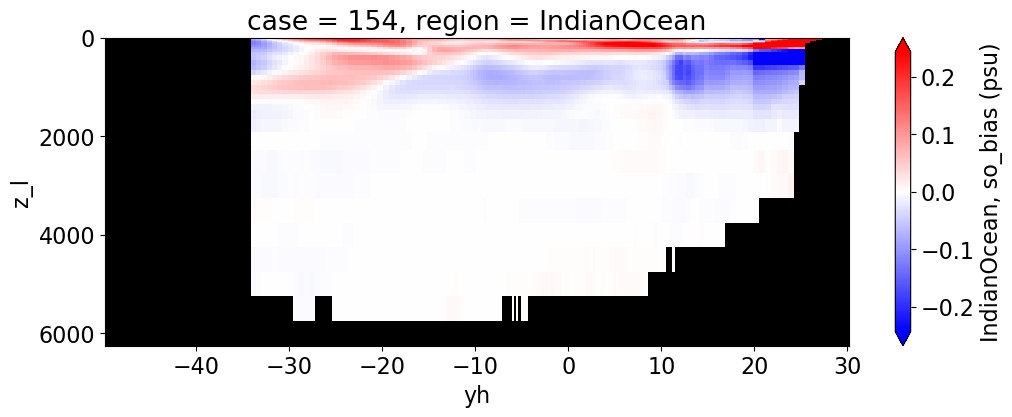
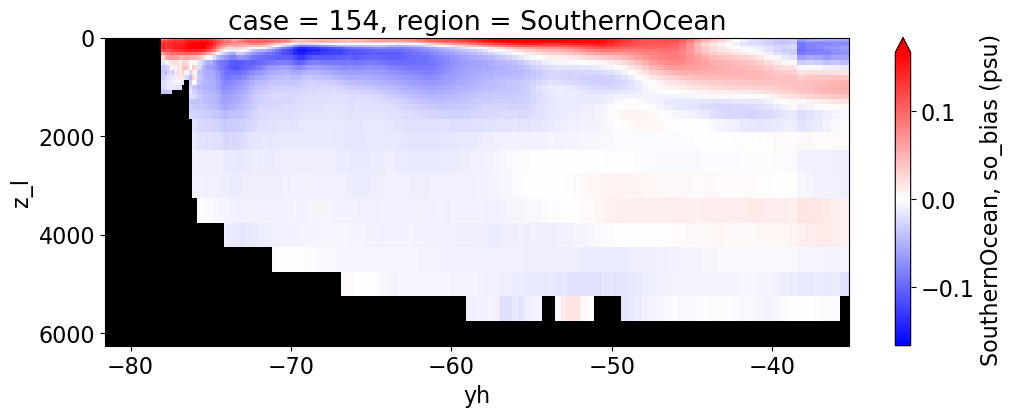
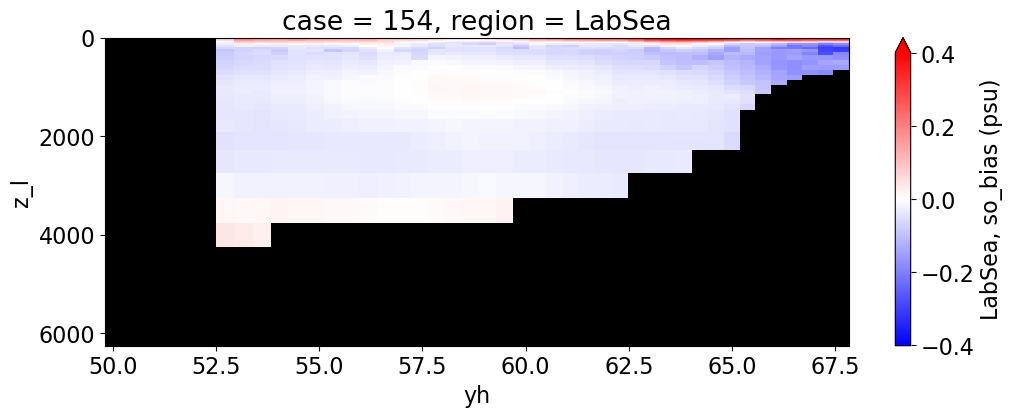
Salinity#
reg = "AtlanticOcean"
var = 'so_bias'
units = 'psu'
regions = ['Global','MedSea','Arctic','PacificOcean',
'AtlanticOcean','IndianOcean','SouthernOcean',
'LabSea']
cmap = plt.get_cmap('bwr')
cmap.set_bad(color='black')
matplotlib.rcParams.update({'font.size': 16})
for reg, j in zip(regions, range(len(regions))):
#print(reg)
da = []
for path, case, i in zip(ocn_path, casename, range(len(casename))):
ds_mom_t = xr.open_dataset(path+case+'_so_time_mean.nc')
atl = basin_code[i].sel(region=reg)
# Set zero values to NaN
atl = atl.where(atl != 0, np.nan)
da.append((ds_mom_t[var] * atl).weighted(grd_xr[i].areacello.fillna(0)).mean('xh').expand_dims(case=[label[i]]).sel(yh=slice(ylim_min[j], ylim_max[j])))
combined_da = xr.concat(da, dim="case")
col = None
if len(combined_da) > 1:
col = "case"
g = combined_da.plot(x="yh", y="z_l", col=col,
yincrease=False, col_wrap=2,
figsize=(12,4), cmap=cmap, robust=True,
cbar_kwargs={"label": reg + ', ' + var + ' ({})'.format(units)})
Meridional biases#
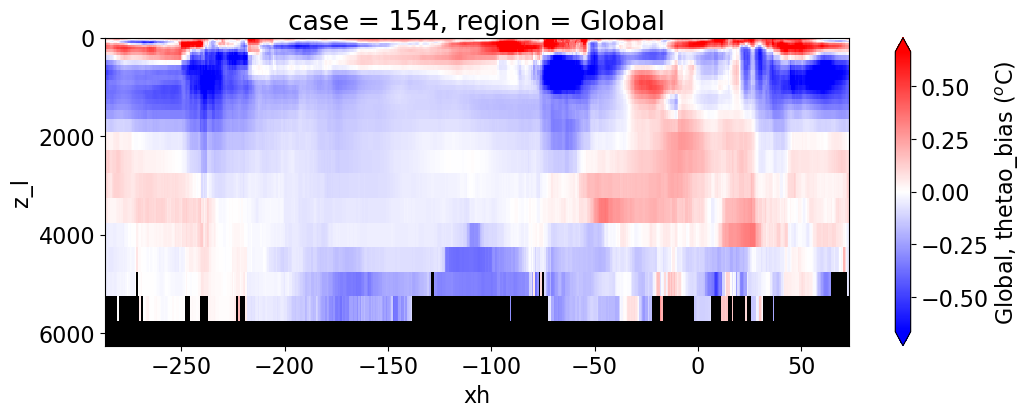
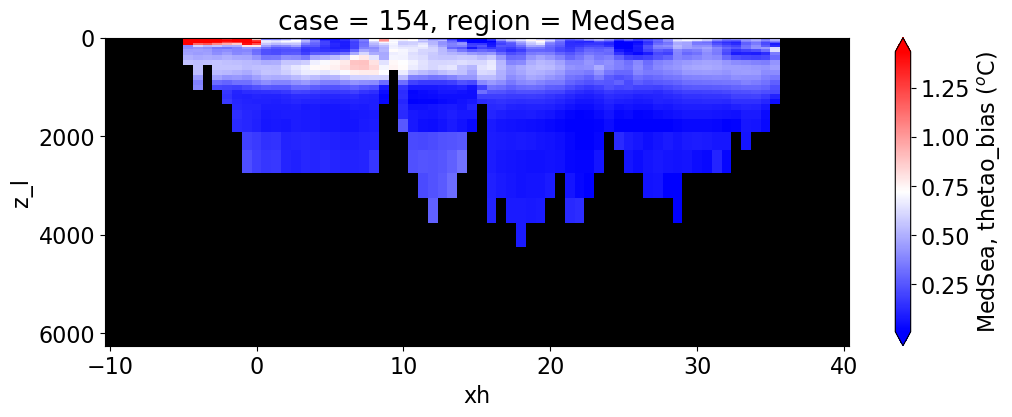
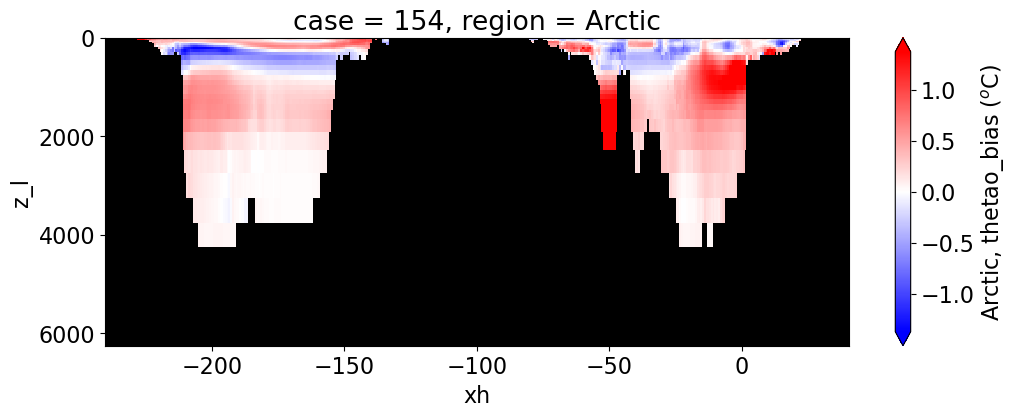
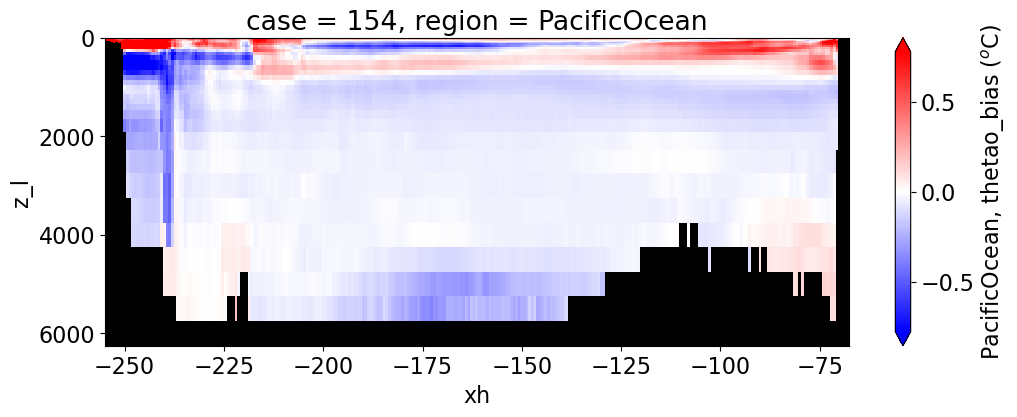
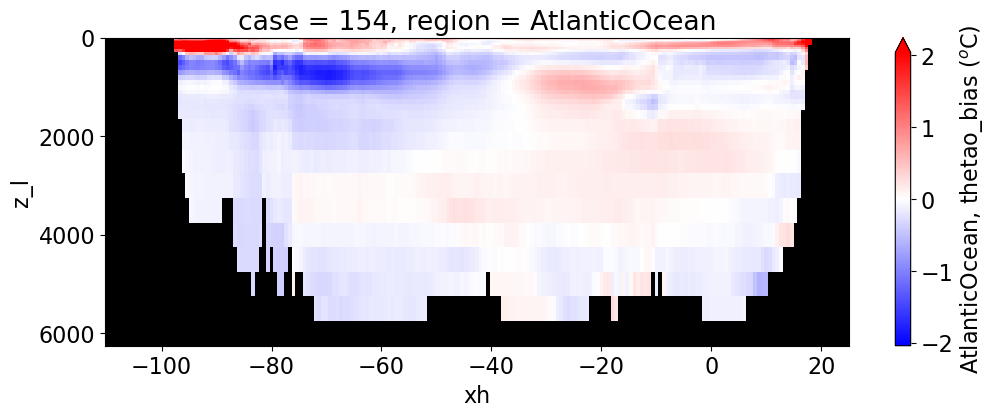
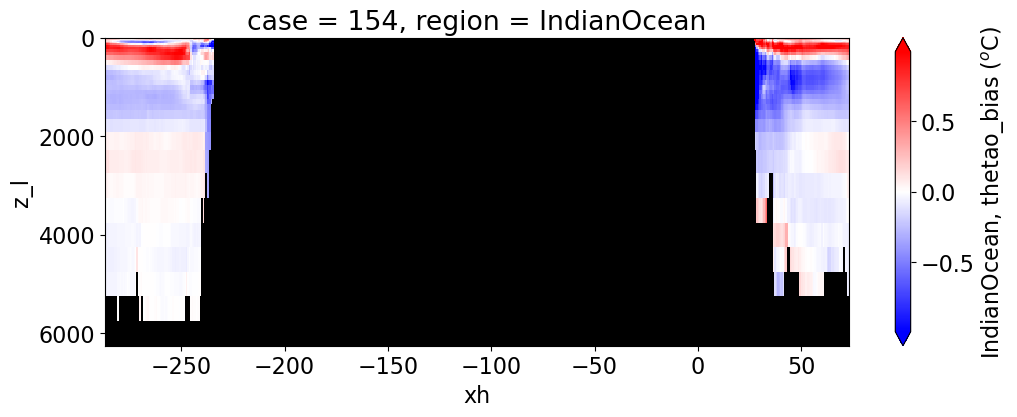
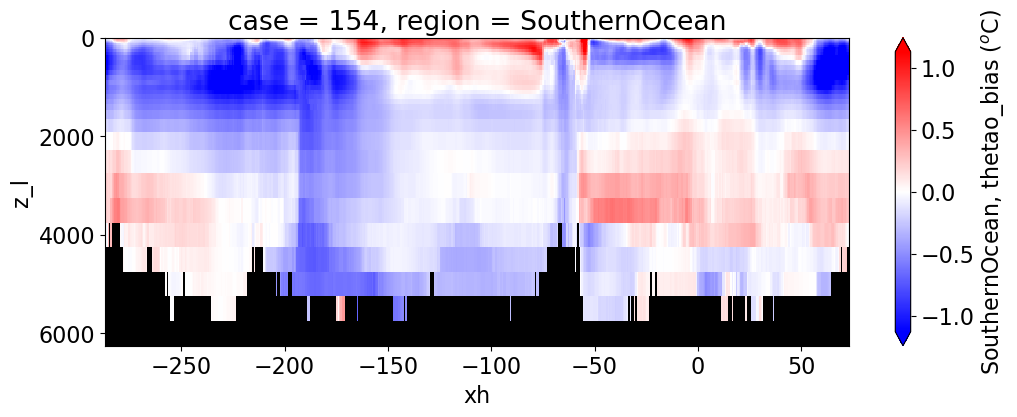
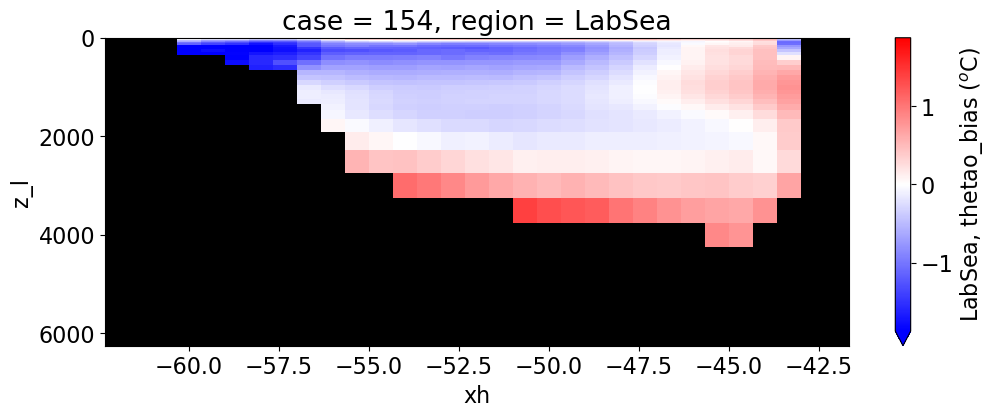
Potential temperature#
reg = "AtlanticOcean"
var = 'thetao_bias'
units = r'$^o$C'
regions = ['Global','MedSea','Arctic','PacificOcean',
'AtlanticOcean','IndianOcean','SouthernOcean',
'LabSea']
xlim_min = [-287, -10, -240, -255, -110, -287, -287, -62]
xlim_max = [ 73, 40, 40, -68, 25, 73, 73, -42 ]
cmap = plt.get_cmap('bwr')
cmap.set_bad(color='black')
matplotlib.rcParams.update({'font.size': 16})
for reg, j in zip(regions, range(len(regions))):
#print(reg)
da = []
for path, case, i in zip(ocn_path, casename, range(len(casename))):
ds_mom_t = xr.open_dataset(path+case+'_thetao_time_mean.nc')
atl = basin_code[i].sel(region=reg)
# Set zero values to NaN
atl = atl.where(atl != 0, np.nan)
da.append((ds_mom_t[var] * \
atl).weighted(grd_xr[i].areacello.fillna(0)).mean('yh').expand_dims(case=[label[i]]).sel(xh=slice(xlim_min[j],
xlim_max[j])))
combined_da = xr.concat(da, dim="case")
col = None
if len(combined_da) > 1:
col = "case"
g = combined_da.plot(x="xh", y="z_l", col=col,
yincrease=False, col_wrap=2,
figsize=(12,4), cmap=cmap, robust=True,
cbar_kwargs={"label": reg + ', ' + var + ' ({})'.format(units)})
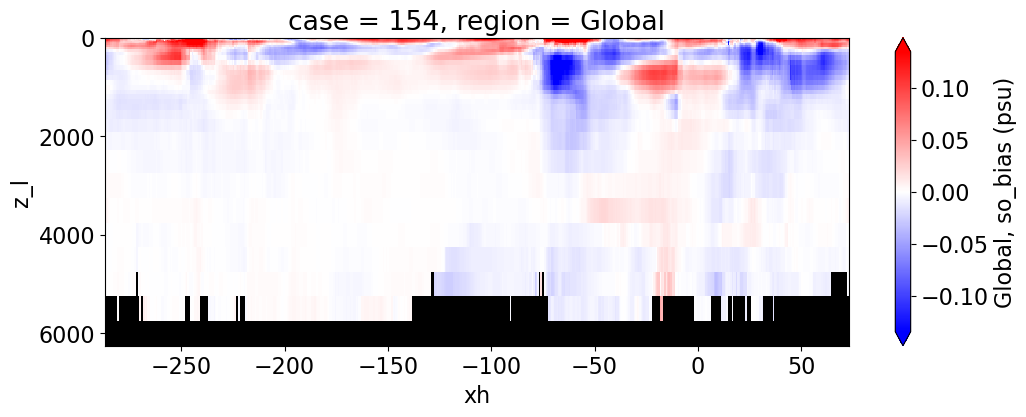
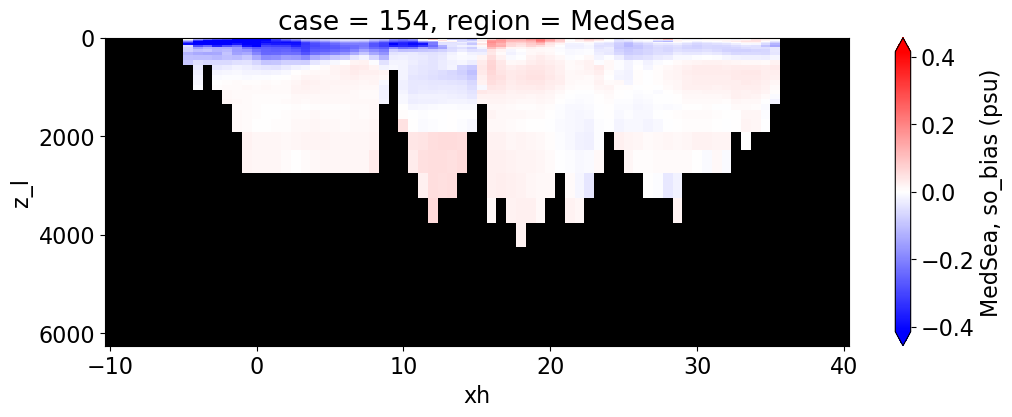
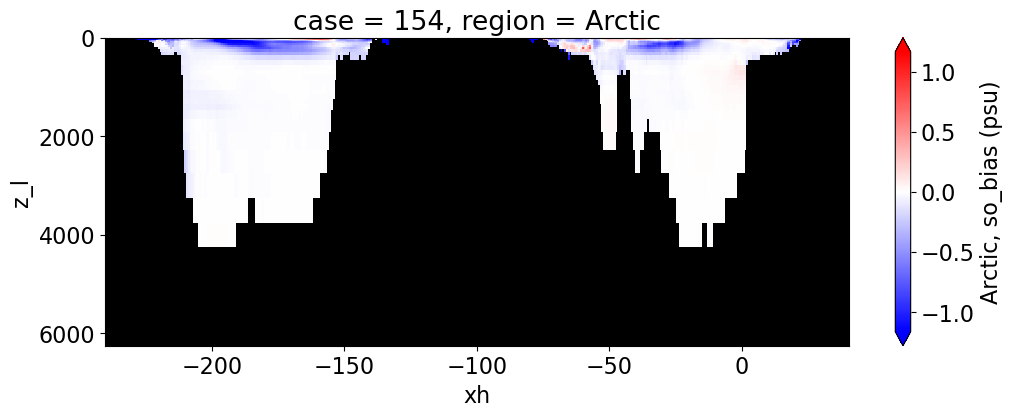
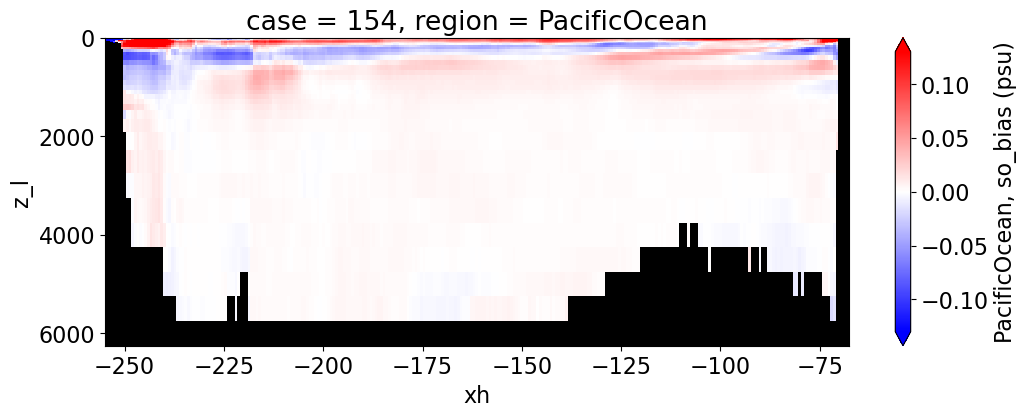
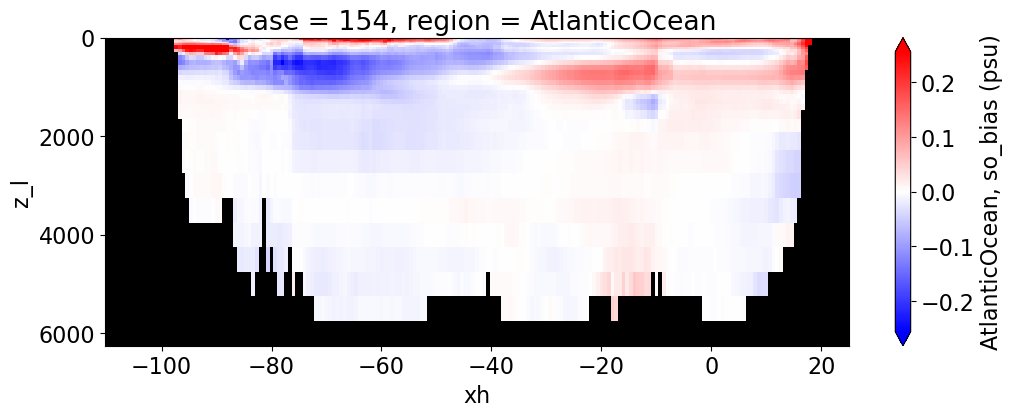
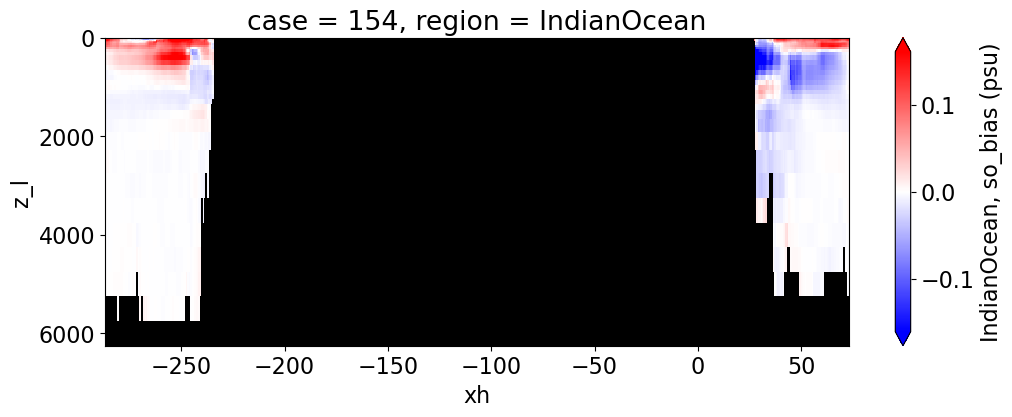
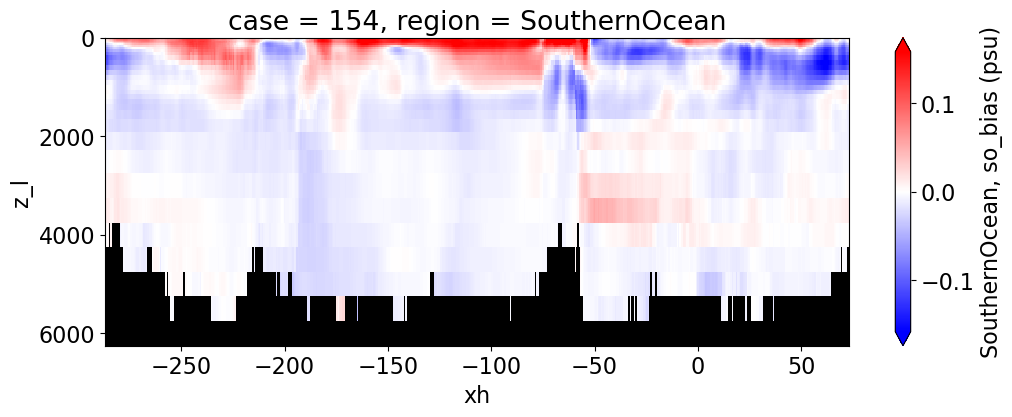
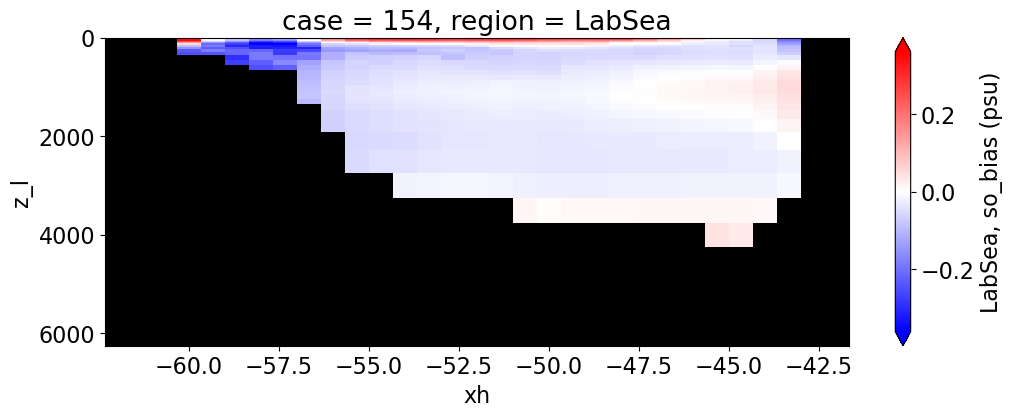
Salinity#
reg = "AtlanticOcean"
var = 'so_bias'
units = 'psu'
regions = ['Global','MedSea','Arctic','PacificOcean',
'AtlanticOcean','IndianOcean','SouthernOcean',
'LabSea']
cmap = plt.get_cmap('bwr')
cmap.set_bad(color='black')
matplotlib.rcParams.update({'font.size': 16})
for reg, j in zip(regions, range(len(regions))):
#print(reg)
da = []
for path, case, i in zip(ocn_path, casename, range(len(casename))):
ds_mom_t = xr.open_dataset(path+case+'_so_time_mean.nc')
atl = basin_code[i].sel(region=reg)
# Set zero values to NaN
atl = atl.where(atl != 0, np.nan)
da.append((ds_mom_t[var] * \
atl).weighted(grd_xr[i].areacello.fillna(0)).mean('yh').expand_dims(case=[label[i]]).sel(xh=slice(xlim_min[j],
xlim_max[j])))
combined_da = xr.concat(da, dim="case")
col = None
if len(combined_da) > 1:
col = "case"
g = combined_da.plot(x="xh", y="z_l", col=col,
yincrease=False, col_wrap=2,
figsize=(12,4), cmap=cmap, robust=True,
cbar_kwargs={"label": reg + ', ' + var + ' ({})'.format(units)})